Jacopo Dapueto
Disentangled representations of microscopy images
Jun 25, 2025Abstract:Microscopy image analysis is fundamental for different applications, from diagnosis to synthetic engineering and environmental monitoring. Modern acquisition systems have granted the possibility to acquire an escalating amount of images, requiring a consequent development of a large collection of deep learning-based automatic image analysis methods. Although deep neural networks have demonstrated great performance in this field, interpretability, an essential requirement for microscopy image analysis, remains an open challenge. This work proposes a Disentangled Representation Learning (DRL) methodology to enhance model interpretability for microscopy image classification. Exploiting benchmark datasets from three different microscopic image domains (plankton, yeast vacuoles, and human cells), we show how a DRL framework, based on transferring a representation learnt from synthetic data, can provide a good trade-off between accuracy and interpretability in this domain.
Transferring disentangled representations: bridging the gap between synthetic and real images
Sep 26, 2024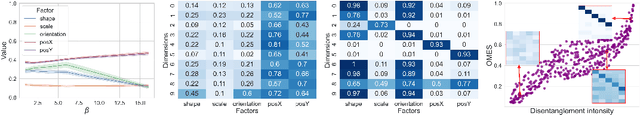
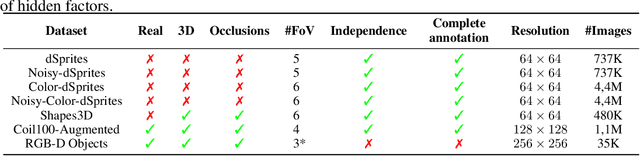
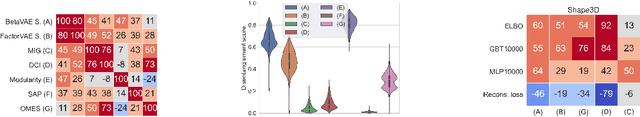
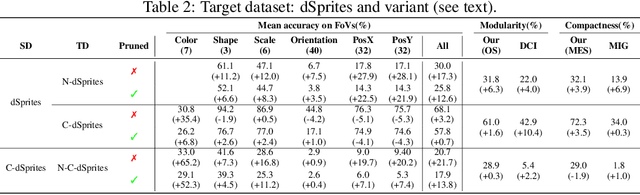
Abstract:Developing meaningful and efficient representations that separate the fundamental structure of the data generation mechanism is crucial in representation learning. However, Disentangled Representation Learning has not fully shown its potential on real images, because of correlated generative factors, their resolution and limited access to ground truth labels. Specifically on the latter, we investigate the possibility of leveraging synthetic data to learn general-purpose disentangled representations applicable to real data, discussing the effect of fine-tuning and what properties of disentanglement are preserved after the transfer. We provide an extensive empirical study to address these issues. In addition, we propose a new interpretable intervention-based metric, to measure the quality of factors encoding in the representation. Our results indicate that some level of disentanglement, transferring a representation from synthetic to real data, is possible and effective.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge