Jörn Schulz
Non-Euclidean Analysis of Joint Variations in Multi-Object Shapes
Sep 07, 2021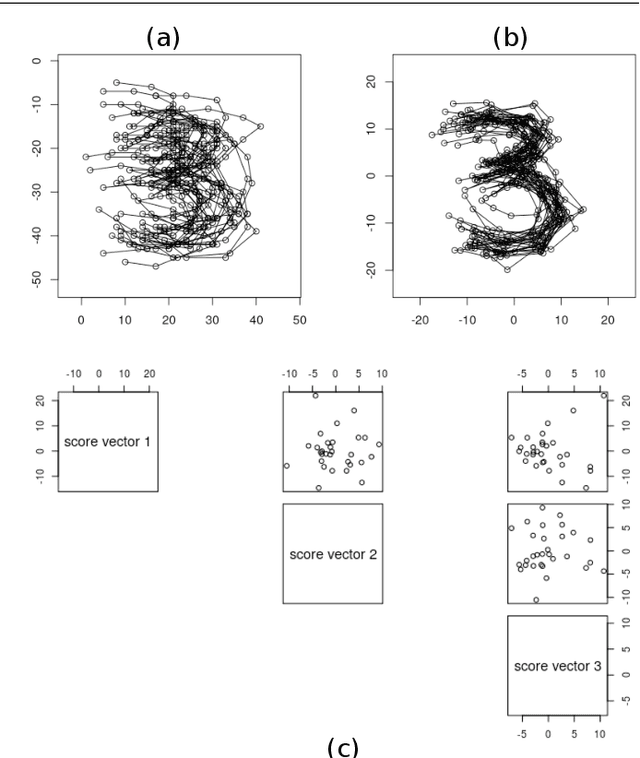
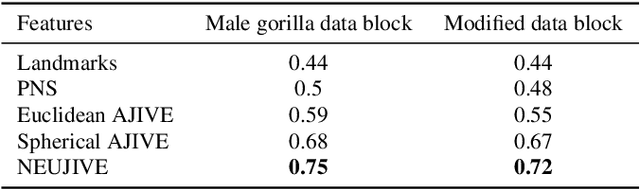
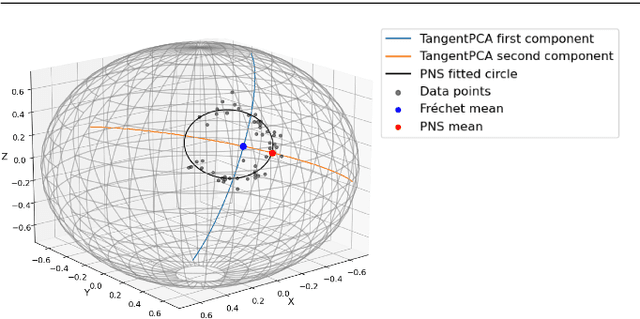
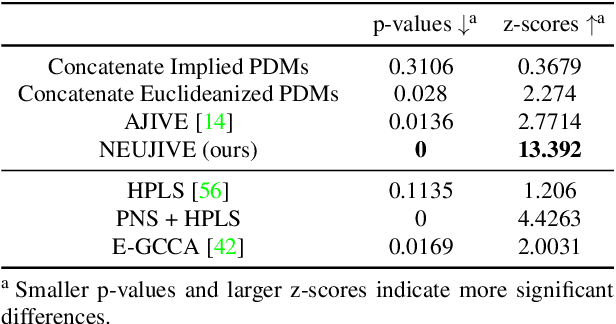
Abstract:This paper considers joint analysis of multiple functionally related structures in classification tasks. In particular, our method developed is driven by how functionally correlated brain structures vary together between autism and control groups. To do so, we devised a method based on a novel combination of (1) non-Euclidean statistics that can faithfully represent non-Euclidean data in Euclidean spaces and (2) a non-parametric integrative analysis method that can decompose multi-block Euclidean data into joint, individual, and residual structures. We find that the resulting joint structure is effective, robust, and interpretable in recognizing the underlying patterns of the joint variation of multi-block non-Euclidean data. We verified the method in classifying the structural shape data collected from cases that developed and did not develop into Autistic Spectrum Disorder (ASD).
Statistical analysis of locally parameterized shapes
Aug 18, 2021



Abstract:The alignment of shapes has been a crucial step in statistical shape analysis, for example, in calculating mean shape, detecting locational differences between two shape populations, and classification. Procrustes alignment is the most commonly used method and state of the art. In this work, we uncover that alignment might seriously affect the statistical analysis. For example, alignment can induce false shape differences and lead to misleading results and interpretations. We propose a novel hierarchical shape parameterization based on local coordinate systems. The local parameterized shapes are translation and rotation invariant. Thus, the inherent alignment problems from the commonly used global coordinate system for shape representation can be avoided using this parameterization. The new parameterization is also superior for shape deformation and simulation. The method's power is demonstrated on the hypothesis testing of simulated data as well as the left hippocampi of patients with Parkinson's disease and controls.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge