Ivan Štajduhar
Building RadiologyNET: Unsupervised annotation of a large-scale multimodal medical database
Jul 27, 2023Abstract:Background and objective: The usage of machine learning in medical diagnosis and treatment has witnessed significant growth in recent years through the development of computer-aided diagnosis systems that are often relying on annotated medical radiology images. However, the availability of large annotated image datasets remains a major obstacle since the process of annotation is time-consuming and costly. This paper explores how to automatically annotate a database of medical radiology images with regard to their semantic similarity. Material and methods: An automated, unsupervised approach is used to construct a large annotated dataset of medical radiology images originating from Clinical Hospital Centre Rijeka, Croatia, utilising multimodal sources, including images, DICOM metadata, and narrative diagnoses. Several appropriate feature extractors are tested for each of the data sources, and their utility is evaluated using k-means and k-medoids clustering on a representative data subset. Results: The optimal feature extractors are then integrated into a multimodal representation, which is then clustered to create an automated pipeline for labelling a precursor dataset of 1,337,926 medical images into 50 clusters of visually similar images. The quality of the clusters is assessed by examining their homogeneity and mutual information, taking into account the anatomical region and modality representation. Conclusion: The results suggest that fusing the embeddings of all three data sources together works best for the task of unsupervised clustering of large-scale medical data, resulting in the most concise clusters. Hence, this work is the first step towards building a much larger and more fine-grained annotated dataset of medical radiology images.
ML-Based Approach for NFL Defensive Pass Interference Prediction Using GPS Tracking Data
Jun 24, 2022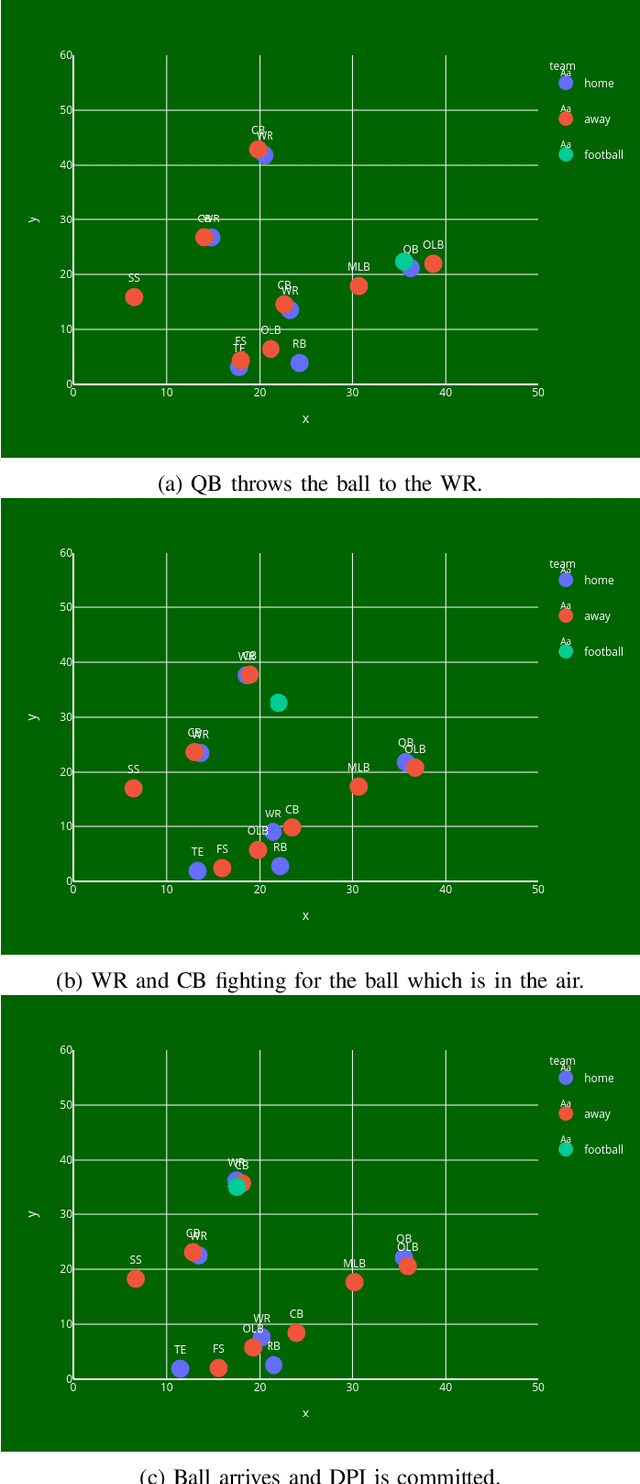
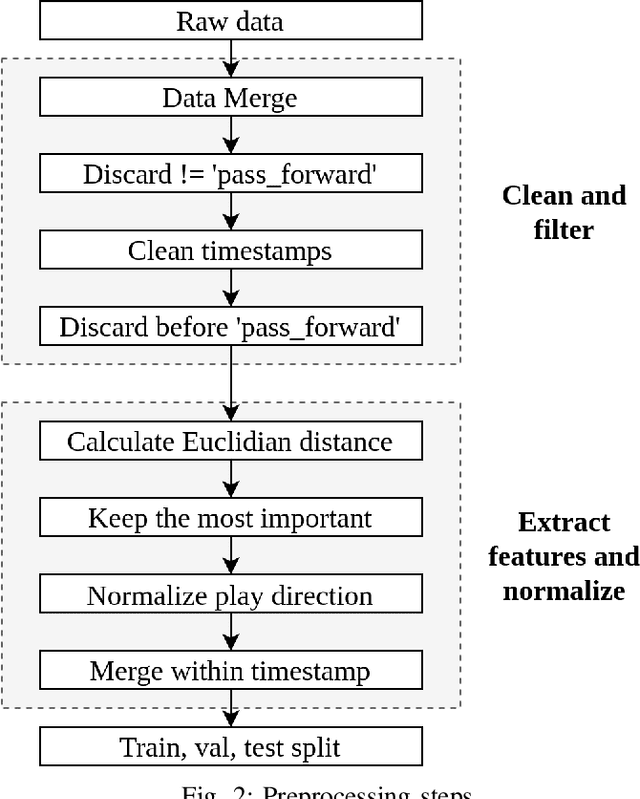


Abstract:Defensive Pass Interference (DPI) is one of the most impactful penalties in the NFL. DPI is a spot foul, yielding an automatic first down to the team in possession. With such an influence on the game, referees have no room for a mistake. It is also a very rare event, which happens 1-2 times per 100 pass attempts. With technology improving and many IoT wearables being put on the athletes to collect valuable data, there is a solid ground for applying machine learning (ML) techniques to improve every aspect of the game. The work presented here is the first attempt in predicting DPI using player tracking GPS data. The data we used was collected by NFL's Next Gen Stats throughout the 2018 regular season. We present ML models for highly imbalanced time-series binary classification: LSTM, GRU, ANN, and Multivariate LSTM-FCN. Results showed that using GPS tracking data to predict DPI has limited success. The best performing models had high recall with low precision which resulted in the classification of many false positive examples. Looking closely at the data confirmed that there is just not enough information to determine whether a foul was committed. This study might serve as a filter for multi-step pipeline for video sequence classification which could be able to solve this problem.
Deep embedded clustering algorithm for clustering PACS repositories
Jun 24, 2022



Abstract:Creating large datasets of medical radiology images from several sources can be challenging because of the differences in the acquisition and storage standards. One possible way of controlling and/or assessing the image selection process is through medical image clustering. This, however, requires an efficient method for learning latent image representations. In this paper, we tackle the problem of fully-unsupervised clustering of medical images using pixel data only. We test the performance of several contemporary approaches, built on top of a convolutional autoencoder (CAE) - convolutional deep embedded clustering (CDEC) and convolutional improved deep embedded clustering (CIDEC) - and three approaches based on preset feature extraction - histogram of oriented gradients (HOG), local binary pattern (LBP) and principal component analysis (PCA). CDEC and CIDEC are end-to-end clustering solutions, involving simultaneous learning of latent representations and clustering assignments, whereas the remaining approaches rely on k-means clustering from fixed embeddings. We train the models on 30,000 images, and test them using a separate test set consisting of 8,000 images. We sampled the data from the PACS repository archive of the Clinical Hospital Centre Rijeka. For evaluation, we use silhouette score, homogeneity score and normalised mutual information (NMI) on two target parameters, closely associated with commonly occurring DICOM tags - Modality and anatomical region (adjusted BodyPartExamined tag). CIDEC attains an NMI score of 0.473 with respect to anatomical region, and CDEC attains an NMI score of 0.645 with respect to the tag Modality - both outperforming other commonly used feature descriptors.
Intra-domain and cross-domain transfer learning for time series data -- How transferable are the features?
Jan 12, 2022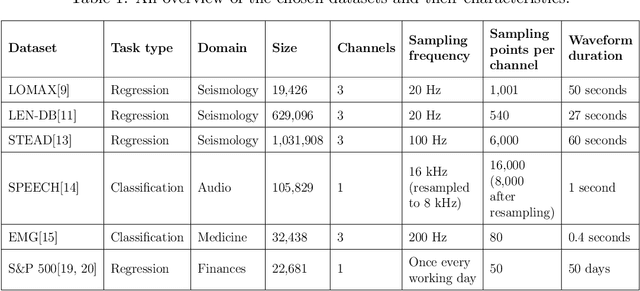
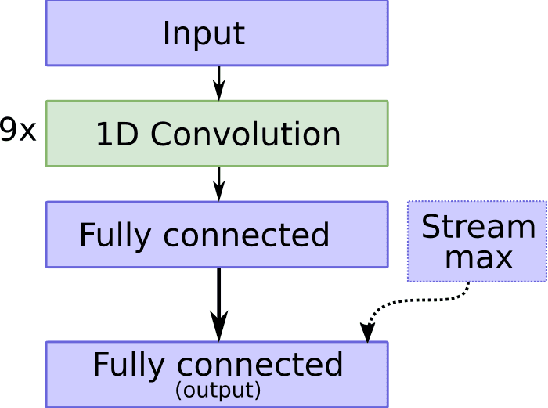
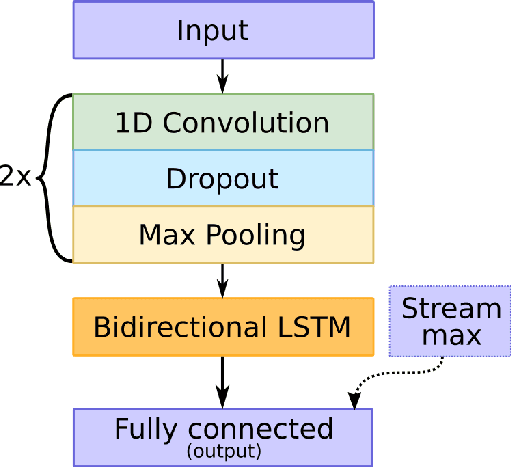
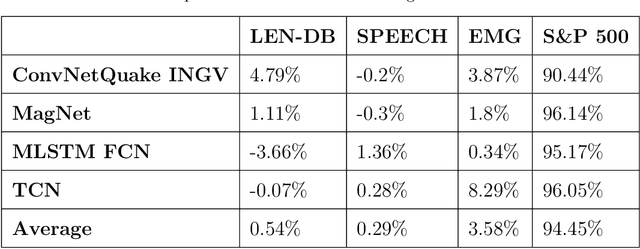
Abstract:In practice, it is very demanding and sometimes impossible to collect datasets of tagged data large enough to successfully train a machine learning model, and one possible solution to this problem is transfer learning. This study aims to assess how transferable are the features between different domains of time series data and under which conditions. The effects of transfer learning are observed in terms of predictive performance of the models and their convergence rate during training. In our experiment, we use reduced data sets of 1,500 and 9,000 data instances to mimic real world conditions. Using the same scaled-down datasets, we trained two sets of machine learning models: those that were trained with transfer learning and those that were trained from scratch. Four machine learning models were used for the experiment. Transfer of knowledge was performed within the same domain of application (seismology), as well as between mutually different domains of application (seismology, speech, medicine, finance). We observe the predictive performance of the models and the convergence rate during the training. In order to confirm the validity of the obtained results, we repeated the experiments seven times and applied statistical tests to confirm the significance of the results. The general conclusion of our study is that transfer learning is very likely to either increase or not negatively affect the predictive performance of the model or its convergence rate. The collected data is analysed in more details to determine which source and target domains are compatible for transfer of knowledge. We also analyse the effect of target dataset size and the selection of model and its hyperparameters on the effects of transfer learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge