Illia Horenko
Fast and close Shannon entropy approximation
May 20, 2025Abstract:Shannon entropy (SE) and its quantum mechanical analogue von Neumann entropy are key components in many tools used in physics, information theory, machine learning (ML) and quantum computing. Besides of the significant amounts of SE computations required in these fields, the singularity of the SE gradient is one of the central mathematical reason inducing the high cost, frequently low robustness and slow convergence of such tools. Here we propose the Fast Entropy Approximation (FEA) - a non-singular rational approximation of Shannon entropy and its gradient that achieves a mean absolute error of $10^{-3}$, which is approximately $20$ times lower than comparable state-of-the-art methods. FEA allows around $50\%$ faster computation, requiring only $5$ to $6$ elementary computational operations, as compared to tens of elementary operations behind the fastest entropy computation algorithms with table look-ups, bitshifts, or series approximations. On a set of common benchmarks for the feature selection problem in machine learning, we show that the combined effect of fewer elementary operations, low approximation error, and a non-singular gradient allows significantly better model quality and enables ML feature extraction that is two to three orders of magnitude faster and computationally cheaper when incorporating FEA into AI tools.
Towards Generalized Entropic Sparsification for Convolutional Neural Networks
Apr 06, 2024



Abstract:Convolutional neural networks (CNNs) are reported to be overparametrized. The search for optimal (minimal) and sufficient architecture is an NP-hard problem as the hyperparameter space for possible network configurations is vast. Here, we introduce a layer-by-layer data-driven pruning method based on the mathematical idea aiming at a computationally-scalable entropic relaxation of the pruning problem. The sparse subnetwork is found from the pre-trained (full) CNN using the network entropy minimization as a sparsity constraint. This allows deploying a numerically scalable algorithm with a sublinear scaling cost. The method is validated on several benchmarks (architectures): (i) MNIST (LeNet) with sparsity 55%-84% and loss in accuracy 0.1%-0.5%, and (ii) CIFAR-10 (VGG-16, ResNet18) with sparsity 73-89% and loss in accuracy 0.1%-0.5%.
Gauge-optimal approximate learning for small data classification problems
Oct 29, 2023Abstract:Small data learning problems are characterized by a significant discrepancy between the limited amount of response variable observations and the large feature space dimension. In this setting, the common learning tools struggle to identify the features important for the classification task from those that bear no relevant information, and cannot derive an appropriate learning rule which allows to discriminate between different classes. As a potential solution to this problem, here we exploit the idea of reducing and rotating the feature space in a lower-dimensional gauge and propose the Gauge-Optimal Approximate Learning (GOAL) algorithm, which provides an analytically tractable joint solution to the dimension reduction, feature segmentation and classification problems for small data learning problems. We prove that the optimal solution of the GOAL algorithm consists in piecewise-linear functions in the Euclidean space, and that it can be approximated through a monotonically convergent algorithm which presents -- under the assumption of a discrete segmentation of the feature space -- a closed-form solution for each optimization substep and an overall linear iteration cost scaling. The GOAL algorithm has been compared to other state-of-the-art machine learning (ML) tools on both synthetic data and challenging real-world applications from climate science and bioinformatics (i.e., prediction of the El Nino Southern Oscillation and inference of epigenetically-induced gene-activity networks from limited experimental data). The experimental results show that the proposed algorithm outperforms the reported best competitors for these problems both in learning performance and computational cost.
On existence, uniqueness and scalability of adversarial robustness measures for AI classifiers
Oct 19, 2023Abstract:Simply-verifiable mathematical conditions for existence, uniqueness and explicit analytical computation of minimal adversarial paths (MAP) and minimal adversarial distances (MAD) for (locally) uniquely-invertible classifiers, for generalized linear models (GLM), and for entropic AI (EAI) are formulated and proven. Practical computation of MAP and MAD, their comparison and interpretations for various classes of AI tools (for neuronal networks, boosted random forests, GLM and EAI) are demonstrated on the common synthetic benchmarks: on a double Swiss roll spiral and its extensions, as well as on the two biomedical data problems (for the health insurance claim predictions, and for the heart attack lethality classification). On biomedical applications it is demonstrated how MAP provides unique minimal patient-specific risk-mitigating interventions in the predefined subsets of accessible control variables.
Linearly-scalable learning of smooth low-dimensional patterns with permutation-aided entropic dimension reduction
Jun 17, 2023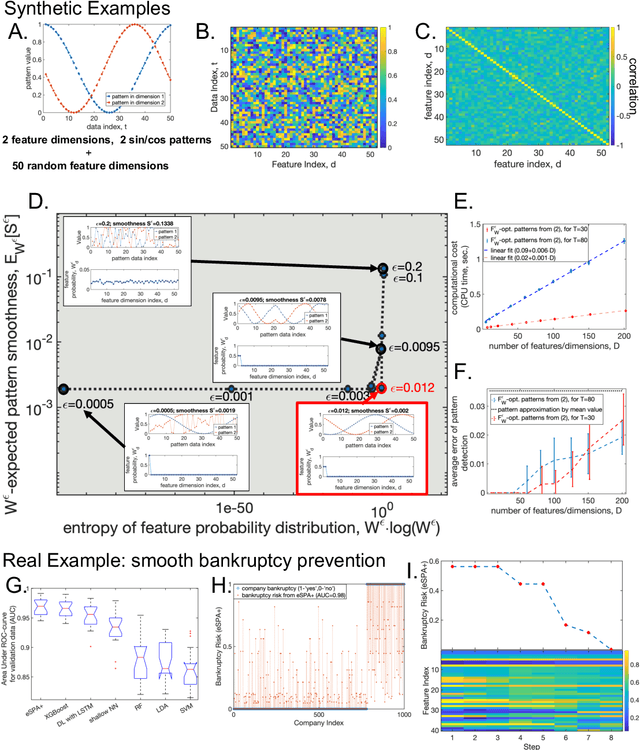
Abstract:In many data science applications, the objective is to extract appropriately-ordered smooth low-dimensional data patterns from high-dimensional data sets. This is challenging since common sorting algorithms are primarily aiming at finding monotonic orderings in low-dimensional data, whereas typical dimension reduction and feature extraction algorithms are not primarily designed for extracting smooth low-dimensional data patterns. We show that when selecting the Euclidean smoothness as a pattern quality criterium, both of these problems (finding the optimal 'crisp' data permutation and extracting the sparse set of permuted low-dimensional smooth patterns) can be efficiently solved numerically as one unsupervised entropy-regularized iterative optimization problem. We formulate and prove the conditions for monotonicity and convergence of this linearly-scalable (in dimension) numerical procedure, with the iteration cost scaling of $\mathcal{O}(DT^2)$, where $T$ is the size of the data statistics and $D$ is a feature space dimension. The efficacy of the proposed method is demonstrated through the examination of synthetic examples as well as a real-world application involving the identification of smooth bankruptcy risk minimizing transition patterns from high-dimensional economical data. The results showcase that the statistical properties of the overall time complexity of the method exhibit linear scaling in the dimensionality $D$ within the specified confidence intervals.
Robust learning of data anomalies with analytically-solvable entropic outlier sparsification
Dec 22, 2021
Abstract:Entropic Outlier Sparsification (EOS) is proposed as a robust computational strategy for the detection of data anomalies in a broad class of learning methods, including the unsupervised problems (like detection of non-Gaussian outliers in mostly-Gaussian data) and in the supervised learning with mislabeled data. EOS dwells on the derived analytic closed-form solution of the (weighted) expected error minimization problem subject to the Shannon entropy regularization. In contrast to common regularization strategies requiring computational costs that scale polynomial with the data dimension, identified closed-form solution is proven to impose additional iteration costs that depend linearly on statistics size and are independent of data dimension. Obtained analytic results also explain why the mixtures of spherically-symmetric Gaussians - used heuristically in many popular data analysis algorithms - represent an optimal choice for the non-parametric probability distributions when working with squared Euclidean distances, combining expected error minimality, maximal entropy/unbiasedness, and a linear cost scaling. The performance of EOS is compared to a range of commonly-used tools on synthetic problems and on partially-mislabeled supervised classification problems from biomedicine.
On a scalable entropic breaching of the overfitting barrier in machine learning
Feb 08, 2020



Abstract:Overfitting and treatment of "small data" are among the most challenging problems in the machine learning (ML), when a relatively small data statistics size $T$ is not enough to provide a robust ML fit for a relatively large data feature dimension $D$. Deploying a massively-parallel ML analysis of generic classification problems for different $D$ and $T$, existence of statistically-significant linear overfitting barriers for common ML methods is demonstrated. For example, these results reveal that for a robust classification of bioinformatics-motivated generic problems with the Long Short-Term Memory deep learning classifier (LSTM) one needs in a best case a statistics $T$ that is at least 13.8 times larger then the feature dimension $D$. It is shown that this overfitting barrier can be breached at a $10^{-12}$ fraction of the computational cost by means of the entropy-optimal Scalable Probabilistic Approximations algorithm (eSPA), performing a joint solution of the entropy-optimal Bayesian network inference and feature space segmentation problems. Application of eSPA to experimental single cell RNA sequencing data exhibits a 30-fold classification performance boost when compared to standard bioinformatics tools - and a 7-fold boost when compared to the deep learning LSTM classifier.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge