Hooman Saadat
BEFUnet: A Hybrid CNN-Transformer Architecture for Precise Medical Image Segmentation
Feb 13, 2024Abstract:The accurate segmentation of medical images is critical for various healthcare applications. Convolutional neural networks (CNNs), especially Fully Convolutional Networks (FCNs) like U-Net, have shown remarkable success in medical image segmentation tasks. However, they have limitations in capturing global context and long-range relations, especially for objects with significant variations in shape, scale, and texture. While transformers have achieved state-of-the-art results in natural language processing and image recognition, they face challenges in medical image segmentation due to image locality and translational invariance issues. To address these challenges, this paper proposes an innovative U-shaped network called BEFUnet, which enhances the fusion of body and edge information for precise medical image segmentation. The BEFUnet comprises three main modules, including a novel Local Cross-Attention Feature (LCAF) fusion module, a novel Double-Level Fusion (DLF) module, and dual-branch encoder. The dual-branch encoder consists of an edge encoder and a body encoder. The edge encoder employs PDC blocks for effective edge information extraction, while the body encoder uses the Swin Transformer to capture semantic information with global attention. The LCAF module efficiently fuses edge and body features by selectively performing local cross-attention on features that are spatially close between the two modalities. This local approach significantly reduces computational complexity compared to global cross-attention while ensuring accurate feature matching. BEFUnet demonstrates superior performance over existing methods across various evaluation metrics on medical image segmentation datasets.
Capturing Local and Global Features in Medical Images by Using Ensemble CNN-Transformer
Nov 03, 2023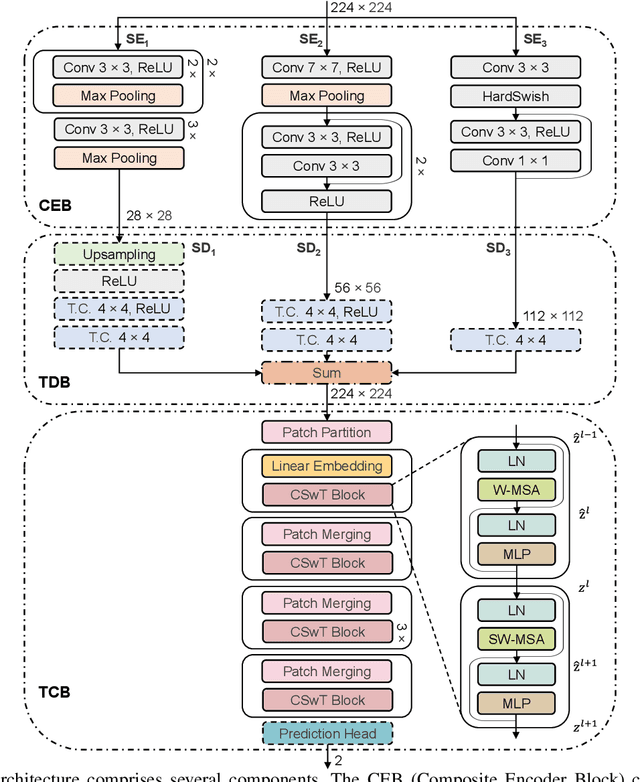
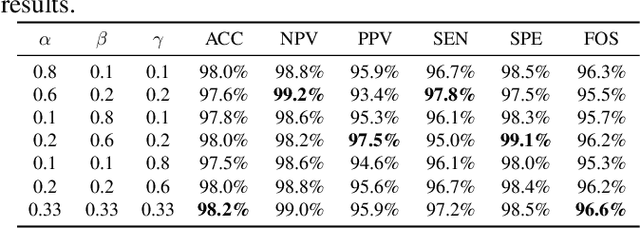
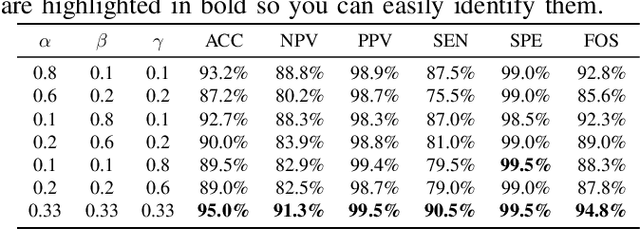
Abstract:This paper introduces a groundbreaking classification model called the Controllable Ensemble Transformer and CNN (CETC) for the analysis of medical images. The CETC model combines the powerful capabilities of convolutional neural networks (CNNs) and transformers to effectively capture both local and global features present in medical images. The model architecture comprises three main components: a convolutional encoder block (CEB), a transposed-convolutional decoder block (TDB), and a transformer classification block (TCB). The CEB is responsible for capturing multi-local features at different scales and draws upon components from VGGNet, ResNet, and MobileNet as backbones. By leveraging this combination, the CEB is able to effectively detect and encode local features. The TDB, on the other hand, consists of sub-decoders that decode and sum the captured features using ensemble coefficients. This enables the model to efficiently integrate the information from multiple scales. Finally, the TCB utilizes the SwT backbone and a specially designed prediction head to capture global features, ensuring a comprehensive understanding of the entire image. The paper provides detailed information on the experimental setup and implementation, including the use of transfer learning, data preprocessing techniques, and training settings. The CETC model is trained and evaluated using two publicly available COVID-19 datasets. Remarkably, the model outperforms existing state-of-the-art models across various evaluation metrics. The experimental results clearly demonstrate the superiority of the CETC model, emphasizing its potential for accurately and efficiently analyzing medical images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge