Hirofumi Kobayashi
Power Stabilization for AI Training Datacenters
Aug 21, 2025Abstract:Large Artificial Intelligence (AI) training workloads spanning several tens of thousands of GPUs present unique power management challenges. These arise due to the high variability in power consumption during the training. Given the synchronous nature of these jobs, during every iteration there is a computation-heavy phase, where each GPU works on the local data, and a communication-heavy phase where all the GPUs synchronize on the data. Because compute-heavy phases require much more power than communication phases, large power swings occur. The amplitude of these power swings is ever increasing with the increase in the size of training jobs. An even bigger challenge arises from the frequency spectrum of these power swings which, if harmonized with critical frequencies of utilities, can cause physical damage to the power grid infrastructure. Therefore, to continue scaling AI training workloads safely, we need to stabilize the power of such workloads. This paper introduces the challenge with production data and explores innovative solutions across the stack: software, GPU hardware, and datacenter infrastructure. We present the pros and cons of each of these approaches and finally present a multi-pronged approach to solving the challenge. The proposed solutions are rigorously tested using a combination of real hardware and Microsoft's in-house cloud power simulator, providing critical insights into the efficacy of these interventions under real-world conditions.
Image Deconvolution via Noise-Tolerant Self-Supervised Inversion
Jun 11, 2020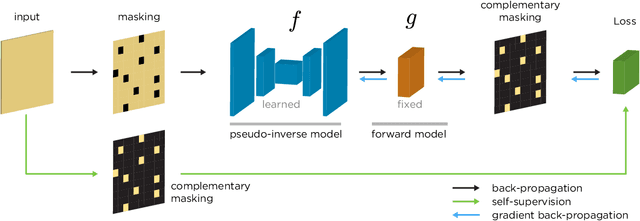
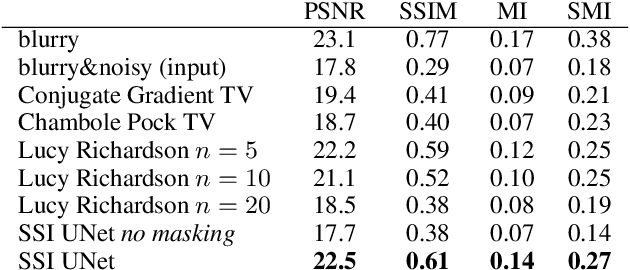
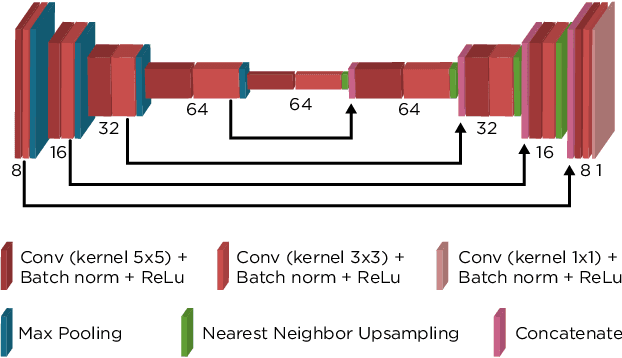

Abstract:We propose a general framework for solving inverse problems in the presence of noise that requires no signal prior, no noise estimate, and no clean training data. We only require that the forward model be available and that the noise be statistically independent across measurement dimensions. We build upon the theory of $\mathcal{J}$-invariant functions (Batson & Royer 2019, arXiv:1901.11365) and show how self-supervised denoising \emph{\`a la} Noise2Self is a special case of learning a noise-tolerant pseudo-inverse of the identity. We demonstrate our approach by showing how a convolutional neural network can be taught in a self-supervised manner to deconvolve images and surpass in image quality classical inversion schemes such as Lucy-Richardson deconvolution.
Modeling the Biological Pathology Continuum with HSIC-regularized Wasserstein Auto-encoders
Jan 20, 2019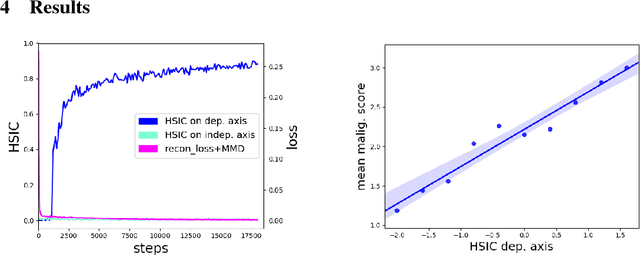
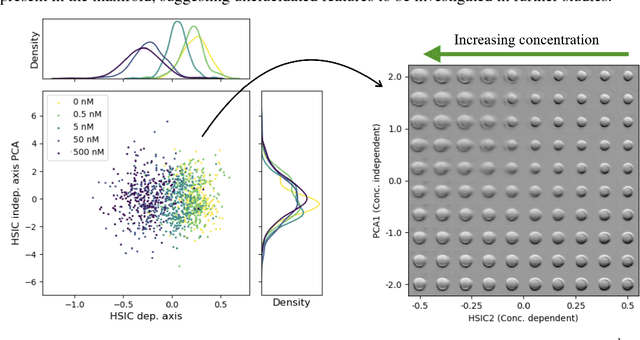
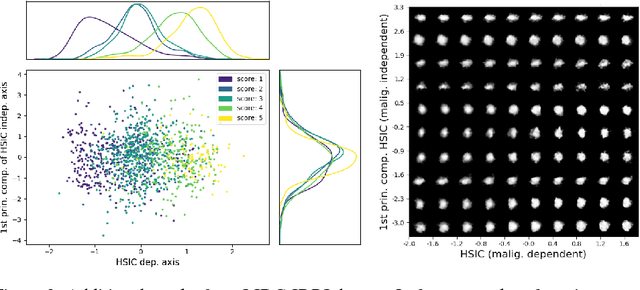
Abstract:A crucial challenge in image-based modeling of biomedical data is to identify trends and features that separate normality and pathology. In many cases, the morphology of the imaged object exhibits continuous change as it deviates from normality, and thus a generative model can be trained to model this morphological continuum. Moreover, given side information that correlates to certain trend in morphological change, a latent variable model can be regularized such that its latent representation reflects this side information. In this work, we use the Wasserstein Auto-encoder to model this pathology continuum, and apply the Hilbert-Schmitt Independence Criterion (HSIC) to enforce dependency between certain latent features and the provided side information. We experimentally show that the model can provide disentangled and interpretable latent representations and also generate a continuum of morphological changes that corresponds to change in the side information.
Exploring the Deep Feature Space of a Cell Classification Neural Network
Nov 15, 2018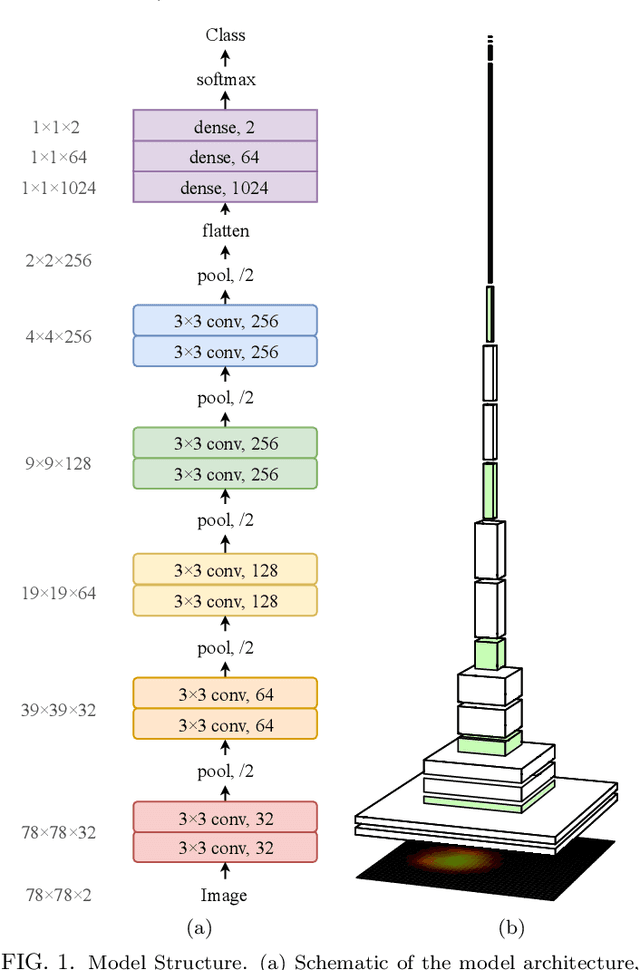
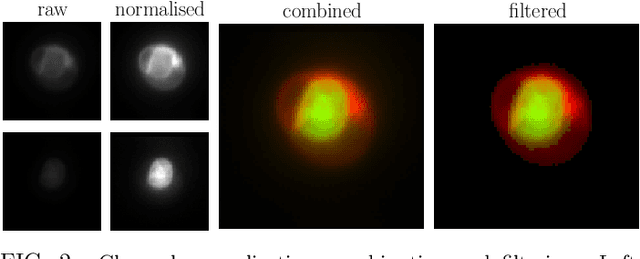
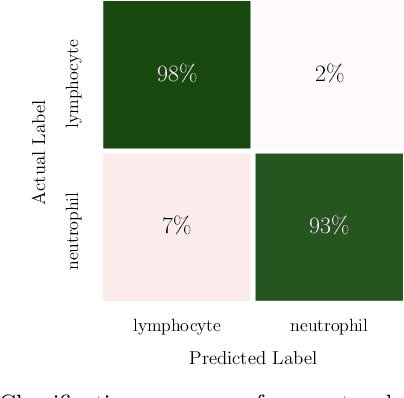
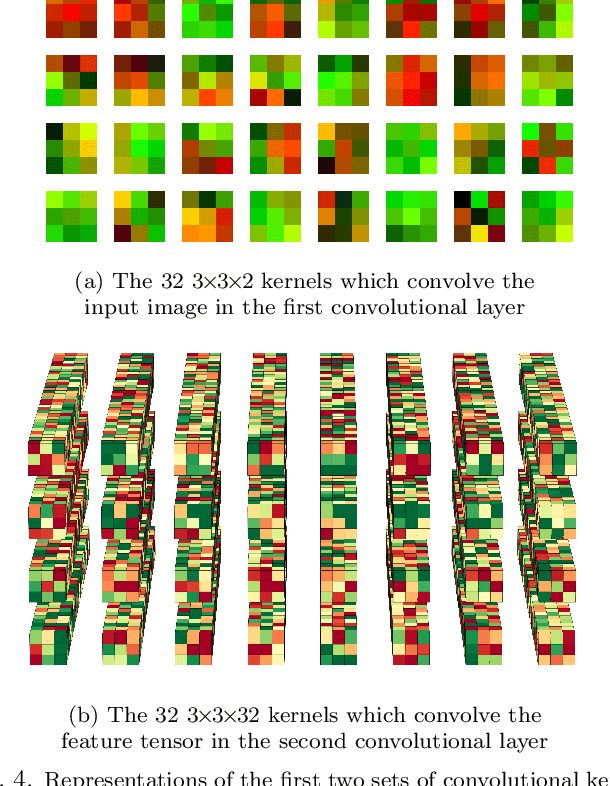
Abstract:In this paper, we present contemporary techniques for visualising the feature space of a deep learning image classification neural network. These techniques are viewed in the context of a feed-forward network trained to classify low resolution fluorescence images of white blood cells captured using optofluidic imaging. The model has two output classes corresponding to two different cell types, which are often difficult to distinguish by eye. This paper has two major sections. The first looks to develop the information space presented by dimension reduction techniques, such as t-SNE, used to embed high-dimensional pre-softmax layer activations into a two-dimensional plane. The second section looks at feature visualisation by optimisation to generate feature images representing the learned features of the network. Using and developing these techniques we visualise class separation and structures within the dataset at various depths using clustering algorithms and feature images; track the development of feature complexity as we ascend the network; and begin to extract the features the network has learnt by modulating single-channel feature images with up-scaled neuron activation maps to distinguish their most salient parts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge