Hassan Ghassemian
Classification of Heart Sounds Using Multi-Branch Deep Convolutional Network and LSTM-CNN
Jul 15, 2024Abstract:This paper presents a fast and cost-effective method for diagnosing cardiac abnormalities with high accuracy and reliability using low-cost systems in clinics. The primary limitation of automatic diagnosing of cardiac diseases is the rarity of correct and acceptable labeled samples, which can be expensive to prepare. To address this issue, two methods are proposed in this work. The first method is a unique Multi-Branch Deep Convolutional Neural Network (MBDCN) architecture inspired by human auditory processing, specifically designed to optimize feature extraction by employing various sizes of convolutional filters and audio signal power spectrum as input. In the second method, called as Long short-term memory-Convolutional Neural (LSCN) model, Additionally, the network architecture includes Long Short-Term Memory (LSTM) network blocks to improve feature extraction in the time domain. The innovative approach of combining multiple parallel branches consisting of the one-dimensional convolutional layers along with LSTM blocks helps in achieving superior results in audio signal processing tasks. The experimental results demonstrate superiority of the proposed methods over the state-of-the-art techniques. The overall classification accuracy of heart sounds with the LSCN network is more than 96%. The efficiency of this network is significant compared to common feature extraction methods such as Mel Frequency Cepstral Coefficients (MFCC) and wavelet transform. Therefore, the proposed method shows promising results in the automatic analysis of heart sounds and has potential applications in the diagnosis and early detection of cardiovascular diseases.
Deep Curriculum Learning for PolSAR Image Classification
Dec 26, 2021
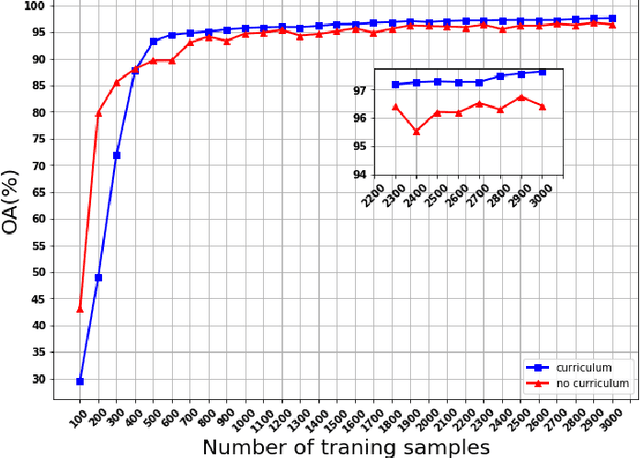
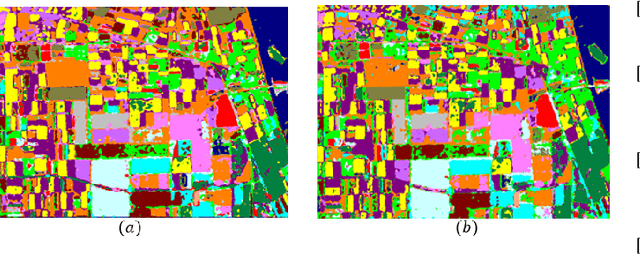
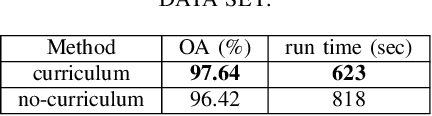
Abstract:Following the great success of curriculum learning in the area of machine learning, a novel deep curriculum learning method proposed in this paper, entitled DCL, particularly for the classification of fully polarimetric synthetic aperture radar (PolSAR) data. This method utilizes the entropy-alpha target decomposition method to estimate the degree of complexity of each PolSAR image patch before applying it to the convolutional neural network (CNN). Also, an accumulative mini-batch pacing function is used to introduce more difficult patches to CNN.Experiments on the widely used data set of AIRSAR Flevoland reveal that the proposed curriculum learning method can not only increase classification accuracy but also lead to faster training convergence.
Multilayer Structured NMF for Spectral Unmixing of Hyperspectral Images
Jun 04, 2015



Abstract:One of the challenges in hyperspectral data analysis is the presence of mixed pixels. Mixed pixels are the result of low spatial resolution of hyperspectral sensors. Spectral unmixing methods decompose a mixed pixel into a set of endmembers and abundance fractions. Due to nonnegativity constraint on abundance fraction values, NMF based methods are well suited to this problem. In this paper multilayer NMF has been used to improve the results of NMF methods for spectral unmixing of hyperspectral data under the linear mixing framework. Sparseness constraint on both spectral signatures and abundance fractions matrices are used in this paper. Evaluation of the proposed algorithm is done using synthetic and real datasets in terms of spectral angle and abundance angle distances. Results show that the proposed algorithm outperforms other previously proposed methods.
Sparsity Constrained Graph Regularized NMF for Spectral Unmixing of Hyperspectral Data
Nov 03, 2014



Abstract:Hyperspectral images contain mixed pixels due to low spatial resolution of hyperspectral sensors. Mixed pixels are pixels containing more than one distinct material called endmembers. The presence percentages of endmembers in mixed pixels are called abundance fractions. Spectral unmixing problem refers to decomposing these pixels into a set of endmembers and abundance fractions. Due to nonnegativity constraint on abundance fractions, nonnegative matrix factorization methods (NMF) have been widely used for solving spectral unmixing problem. In this paper we have used graph regularized NMF (GNMF) method combined with sparseness constraint to decompose mixed pixels in hyperspectral imagery. This method preserves the geometrical structure of data while representing it in low dimensional space. Adaptive regularization parameter based on temperature schedule in simulated annealing method also has been used in this paper for the sparseness term. Proposed algorithm is applied on synthetic and real datasets. Synthetic data is generated based on endmembers from USGS spectral library. AVIRIS Cuprite dataset is used as real dataset for evaluation of proposed method. Results are quantified based on spectral angle distance (SAD) and abundance angle distance (AAD) measures. Results in comparison with other methods show that the proposed method can unmix data more effectively. Specifically for the Cuprite dataset, performance of the proposed method is approximately 10% better than the VCA and Sparse NMF in terms of root mean square of SAD.
Spectral Unmixing of Hyperspectral Imagery using Multilayer NMF
Aug 12, 2014



Abstract:Hyperspectral images contain mixed pixels due to low spatial resolution of hyperspectral sensors. Spectral unmixing problem refers to decomposing mixed pixels into a set of endmembers and abundance fractions. Due to nonnegativity constraint on abundance fractions, nonnegative matrix factorization (NMF) methods have been widely used for solving spectral unmixing problem. In this letter we proposed using multilayer NMF (MLNMF) for the purpose of hyperspectral unmixing. In this approach, spectral signature matrix can be modeled as a product of sparse matrices. In fact MLNMF decomposes the observation matrix iteratively in a number of layers. In each layer, we applied sparseness constraint on spectral signature matrix as well as on abundance fractions matrix. In this way signatures matrix can be sparsely decomposed despite the fact that it is not generally a sparse matrix. The proposed algorithm is applied on synthetic and real datasets. Synthetic data is generated based on endmembers from USGS spectral library. AVIRIS Cuprite dataset has been used as a real dataset for evaluation of proposed method. Results of experiments are quantified based on SAD and AAD measures. Results in comparison with previously proposed methods show that the multilayer approach can unmix data more effectively.
Fusion of Hyperspectral and Panchromatic Images using Spectral Uumixing Results
Oct 22, 2013



Abstract:Hyperspectral imaging, due to providing high spectral resolution images, is one of the most important tools in the remote sensing field. Because of technological restrictions hyperspectral sensors has a limited spatial resolution. On the other hand panchromatic image has a better spatial resolution. Combining this information together can provide a better understanding of the target scene. Spectral unmixing of mixed pixels in hyperspectral images results in spectral signature and abundance fractions of endmembers but gives no information about their location in a mixed pixel. In this paper we have used spectral unmixing results of hyperspectral images and segmentation results of panchromatic image for data fusion. The proposed method has been applied on simulated data using AVRIS Indian Pines datasets. Results show that this method can effectively combine information in hyperspectral and panchromatic images.
Hyperspectral Data Unmixing Using GNMF Method and Sparseness Constraint
Jun 29, 2013



Abstract:Hyperspectral images contain mixed pixels due to low spatial resolution of hyperspectral sensors. Mixed pixels are pixels containing more than one distinct material called endmembers. The presence percentages of endmembers in mixed pixels are called abundance fractions. Spectral unmixing problem refers to decomposing these pixels into a set of endmembers and abundance fractions. Due to nonnegativity constraint on abundance fractions, nonnegative matrix factorization methods (NMF) have been widely used for solving spectral unmixing problem. In this paper we have used graph regularized (GNMF) method with sparseness constraint to unmix hyperspectral data. This method applied on simulated data using AVIRIS Indian Pines dataset and USGS library and results are quantified based on AAD and SAD measures. Results in comparison with other methods show that the proposed method can unmix data more effectively.
Unmixing of Hyperspectral Data Using Robust Statistics-based NMF
Dec 04, 2012



Abstract:Mixed pixels are presented in hyperspectral images due to low spatial resolution of hyperspectral sensors. Spectral unmixing decomposes mixed pixels spectra into endmembers spectra and abundance fractions. In this paper using of robust statistics-based nonnegative matrix factorization (RNMF) for spectral unmixing of hyperspectral data is investigated. RNMF uses a robust cost function and iterative updating procedure, so is not sensitive to outliers. This method has been applied to simulated data using USGS spectral library, AVIRIS and ROSIS datasets. Unmixing results are compared to traditional NMF method based on SAD and AAD measures. Results demonstrate that this method can be used efficiently for hyperspectral unmixing purposes.
Graph Regularized Nonnegative Matrix Factorization for Hyperspectral Data Unmixing
Nov 03, 2011



Abstract:Spectral unmixing is an important tool in hyperspectral data analysis for estimating endmembers and abundance fractions in a mixed pixel. This paper examines the applicability of a recently developed algorithm called graph regularized nonnegative matrix factorization (GNMF) for this aim. The proposed approach exploits the intrinsic geometrical structure of the data besides considering positivity and full additivity constraints. Simulated data based on the measured spectral signatures, is used for evaluating the proposed algorithm. Results in terms of abundance angle distance (AAD) and spectral angle distance (SAD) show that this method can effectively unmix hyperspectral data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge