Haseeb Nazki
MultiPathGAN: Structure Preserving Stain Normalization using Unsupervised Multi-domain Adversarial Network with Perception Loss
Apr 20, 2022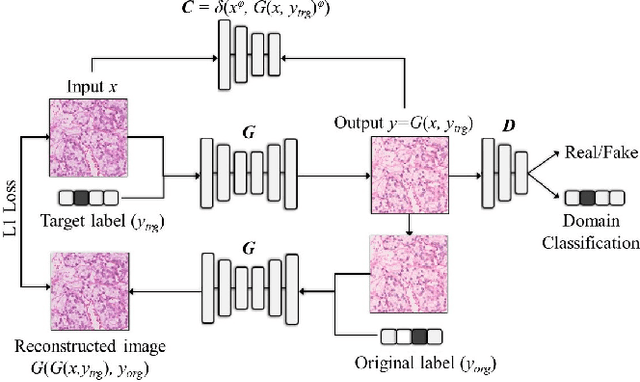
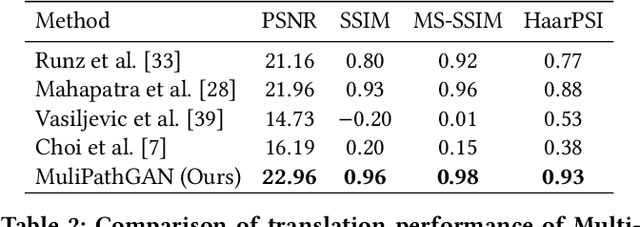
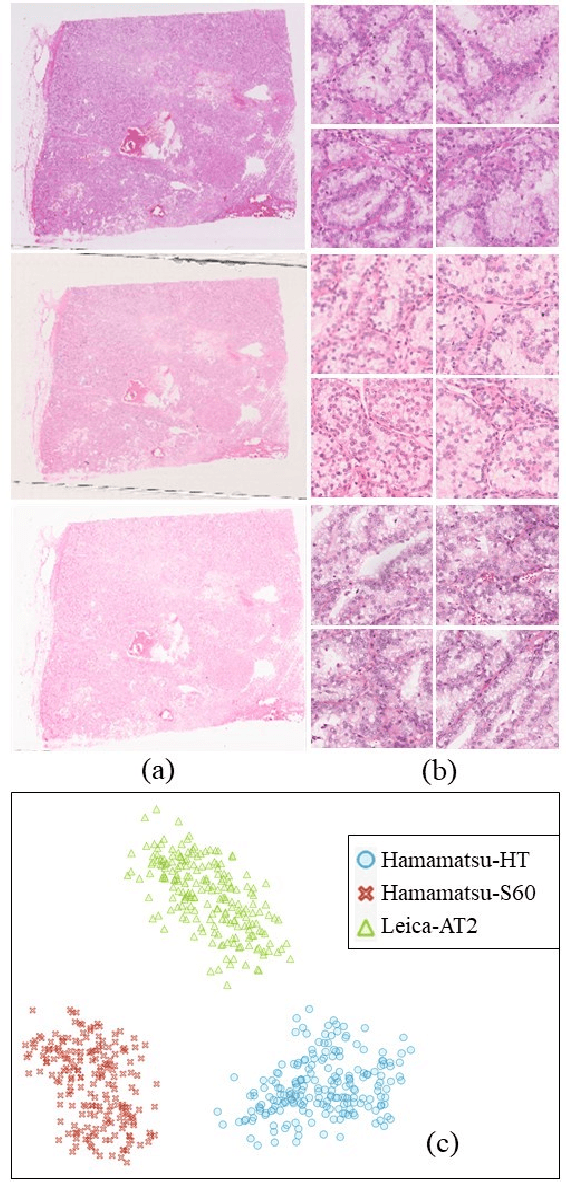
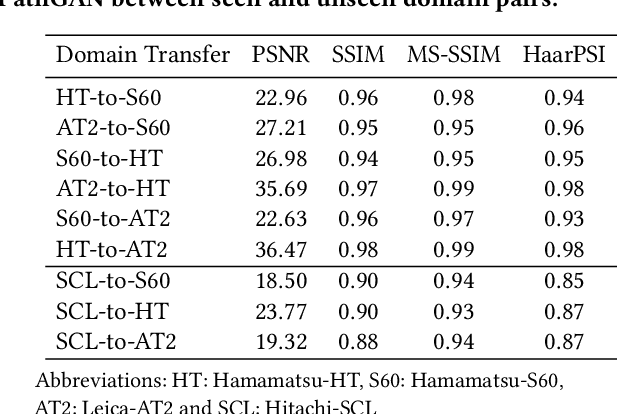
Abstract:Histopathology relies on the analysis of microscopic tissue images to diagnose disease. A crucial part of tissue preparation is staining whereby a dye is used to make the salient tissue components more distinguishable. However, differences in laboratory protocols and scanning devices result in significant confounding appearance variation in the corresponding images. This variation increases both human error and the inter-rater variability, as well as hinders the performance of automatic or semi-automatic methods. In the present paper we introduce an unsupervised adversarial network to translate (and hence normalize) whole slide images across multiple data acquisition domains. Our key contributions are: (i) an adversarial architecture which learns across multiple domains with a single generator-discriminator network using an information flow branch which optimizes for perceptual loss, and (ii) the inclusion of an additional feature extraction network during training which guides the transformation network to keep all the structural features in the tissue image intact. We: (i) demonstrate the effectiveness of the proposed method firstly on H\&E slides of 120 cases of kidney cancer, as well as (ii) show the benefits of the approach on more general problems, such as flexible illumination based natural image enhancement and light source adaptation.
Unsupervised Image Translation using Adversarial Networks for Improved Plant Disease Recognition
Sep 26, 2019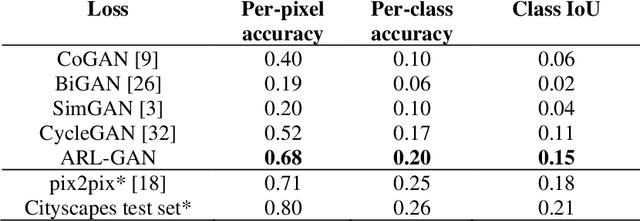
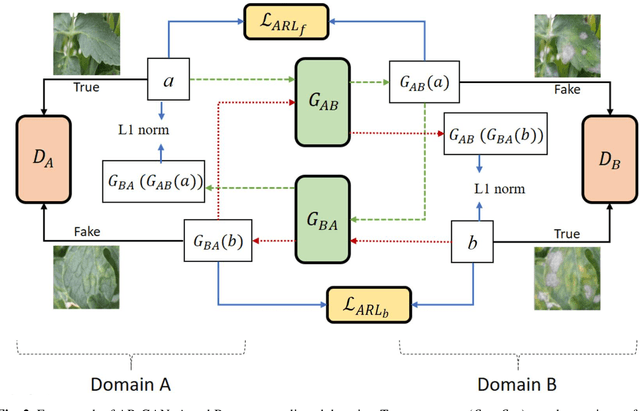
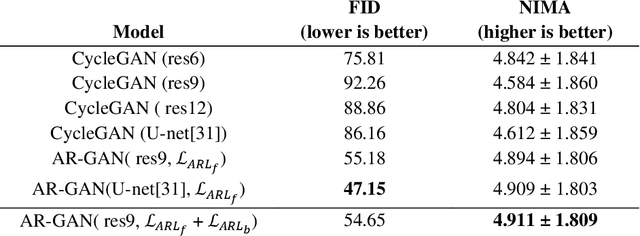
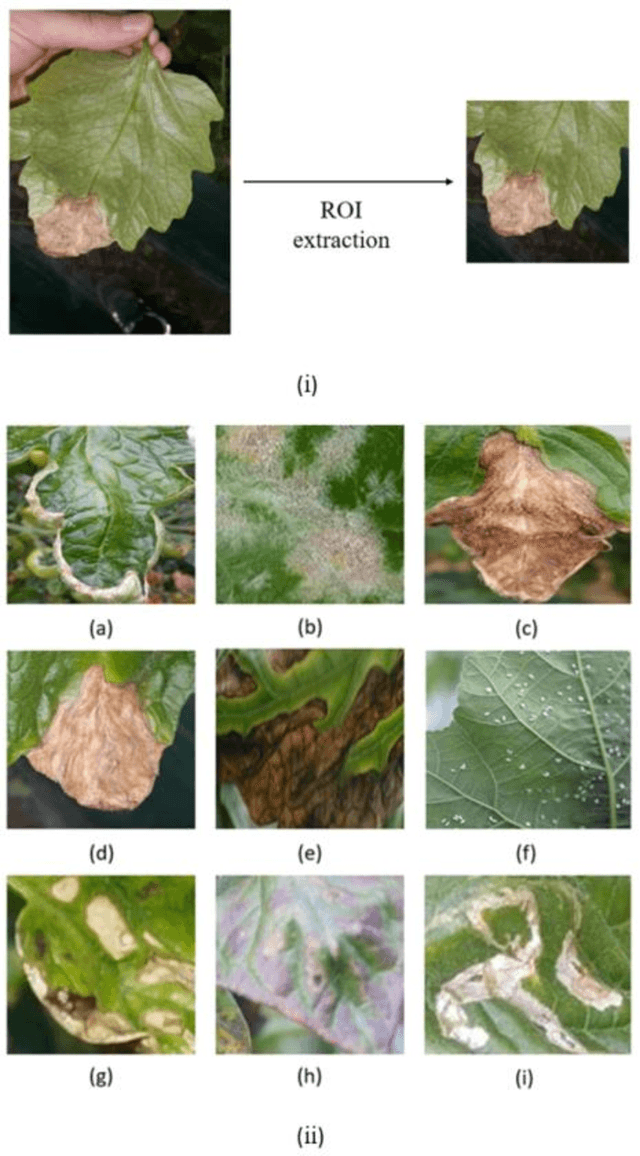
Abstract:Acquisition of data in task-specific applications of machine learning like plant disease recognition is a costly endeavor owing to the requirements of professional human diligence and time constraints. In this paper, we present a simple pipeline that uses GANs in an unsupervised image translation environment to improve learning with respect to the data distribution in a plant disease dataset, reducing the partiality introduced by acute class imbalance and hence shifting the classification decision boundary towards better performance. The empirical analysis of our method is demonstrated on a limited dataset of 2789 tomato plant disease images, highly corrupted with an imbalance in the 9 disease categories. First, we extend the state of the art for the GAN-based image-to-image translation method by enhancing the perceptual quality of the generated images and preserving the semantics. We introduce AR-GAN, where in addition to the adversarial loss, our synthetic image generator optimizes on Activation Reconstruction loss (ARL) function that optimizes feature activations against the natural image. We present visually more compelling synthetic images in comparison to most prominent existing models and evaluate the performance of our GAN framework in terms of various datasets and metrics. Second, we evaluate the performance of a baseline convolutional neural network classifier for improved recognition using the resulting synthetic samples to augment our training set and compare it with the classical data augmentation scheme. We observe a significant improvement in classification accuracy (+5.2%) using generated synthetic samples as compared to (+0.8%) increase using classic augmentation in an equal class distribution environment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge