Grzegorz Bauman
Department of Radiology, Division of Radiological Physics, University Hospital Basel, University of Basel, Basel, Switzerland, Department of Biomedical Engineering, University of Basel, Basel, Switzerland
MRI lung lobe segmentation in pediatric cystic fibrosis patients using a recurrent neural network trained with publicly accessible CT datasets
Aug 31, 2021



Abstract:Purpose: To introduce a widely applicable workflow for pulmonary lobe segmentation of MR images using a recurrent neural network (RNN) trained with chest computed tomography (CT) datasets. The feasibility is demonstrated for 2D coronal ultra-fast balanced steady-state free precession (ufSSFP) MRI. Methods: Lung lobes of 250 publicly accessible CT datasets of adults were segmented with an open-source CT-specific algorithm. To match 2D ufSSFP MRI data of pediatric patients, both CT data and segmentations were translated into pseudo-MR images, masked to suppress anatomy outside the lung. Network-1 was trained with pseudo-MR images and lobe segmentations, and applied to 1000 masked ufSSFP images to predict lobe segmentations. These outputs were directly used as targets to train Network-2 and Network-3 with non-masked ufSSFP data as inputs, and an additional whole-lung mask as input for Network-2. Network predictions were compared to reference manual lobe segmentations of ufSSFP data in twenty pediatric cystic fibrosis patients. Manual lobe segmentations were performed by splitting available whole-lung segmentations into lobes. Results: Network-1 was able to segment the lobes of ufSSFP images, and Network-2 and Network-3 further increased segmentation accuracy and robustness. The average all-lobe Dice similarity coefficients were 95.0$\pm$2.3 (mean$\pm$pooled SD [%]), 96.4$\pm$1.2, 93.0$\pm$1.8, and the average median Hausdorff distances were 6.1$\pm$0.9 (mean$\pm$SD [mm]), 5.3$\pm$1.1, 7.1$\pm$1.3, for Network-1, Network-2, and Network-3, respectively. Conclusions: RNN lung lobe segmentation of 2D ufSSFP imaging is feasible, in good agreement with manual segmentations. The proposed workflow might provide rapid access to automated lobe segmentations for various lung MRI examinations and quantitative analyses.
Recurrent Registration Neural Networks for Deformable Image Registration
Jun 07, 2019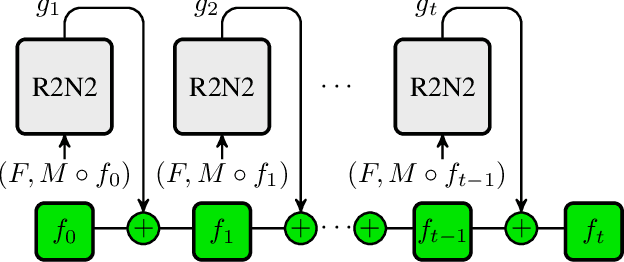
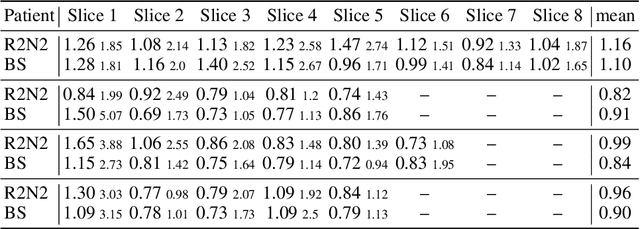
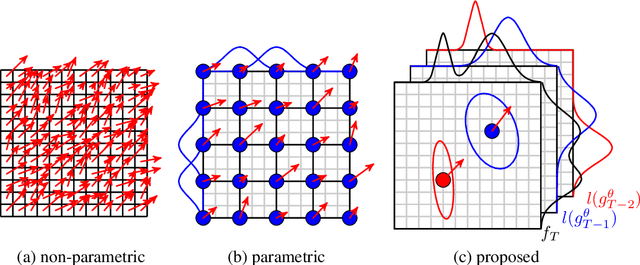
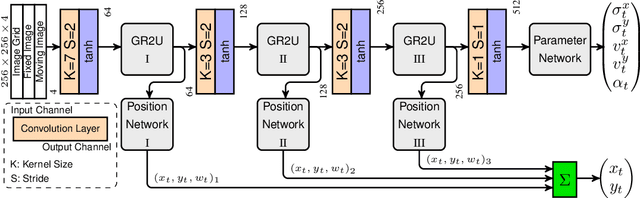
Abstract:Parametric spatial transformation models have been successfully applied to image registration tasks. In such models, the transformation of interest is parameterized by a fixed set of basis functions as for example B-splines. Each basis function is located on a fixed regular grid position among the image domain, because the transformation of interest is not known in advance. As a consequence, not all basis functions will necessarily contribute to the final transformation which results in a non-compact representation of the transformation. We reformulate the pairwise registration problem as a recursive sequence of successive alignments. For each element in the sequence, a local deformation defined by its position, shape, and weight is computed by our recurrent registration neural network. The sum of all local deformations yield the final spatial alignment of both images. Formulating the registration problem in this way allows the network to detect non-aligned regions in the images and to learn how to locally refine the registration properly. In contrast to current non-sequence-based registration methods, our approach iteratively applies local spatial deformations to the images until the desired registration accuracy is achieved. We trained our network on 2D magnetic resonance images of the lung and compared our method to a standard parametric B-spline registration. The experiments show, that our method performs on par for the accuracy but yields a more compact representation of the transformation. Furthermore, we achieve a speedup of around 15 compared to the B-spline registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge