Graham Knott
Deep Active Surface Models
Nov 20, 2020



Abstract:Active Surface Models have a long history of being useful to model complex 3D surfaces but only Active Contours have been used in conjunction with deep networks, and then only to produce the data term as well as meta-parameter maps controlling them. In this paper, we advocate a much tighter integration. We introduce layers that implement them that can be integrated seamlessly into Graph Convolutional Networks to enforce sophisticated smoothness priors at an acceptable computational cost. We will show that the resulting Deep Active Surface Models outperform equivalent architectures that use traditional regularization loss terms to impose smoothness priors for 3D surface reconstruction from 2D images and for 3D volume segmentation.
VM-Net: Mesh Modeling to Assist Segmentation in Volumetric Data
Dec 08, 2019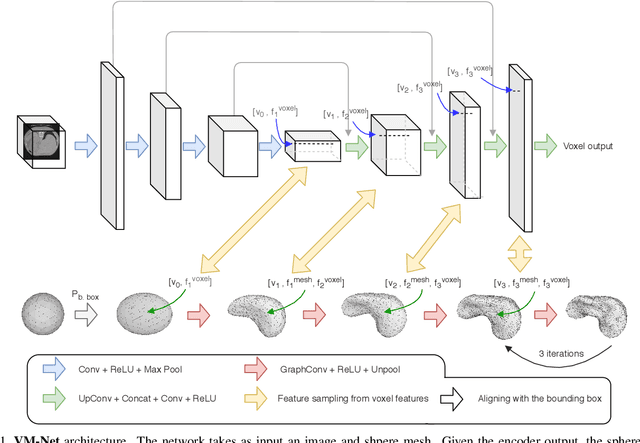
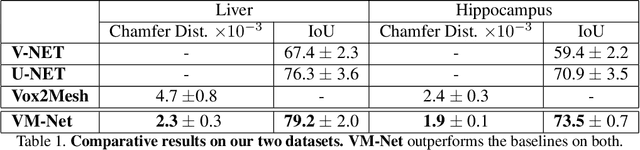
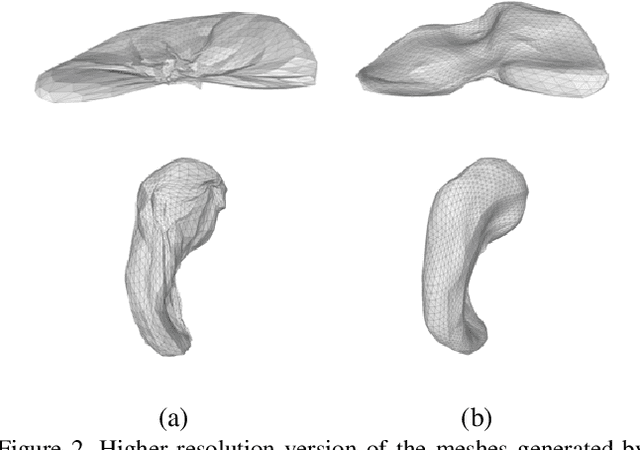
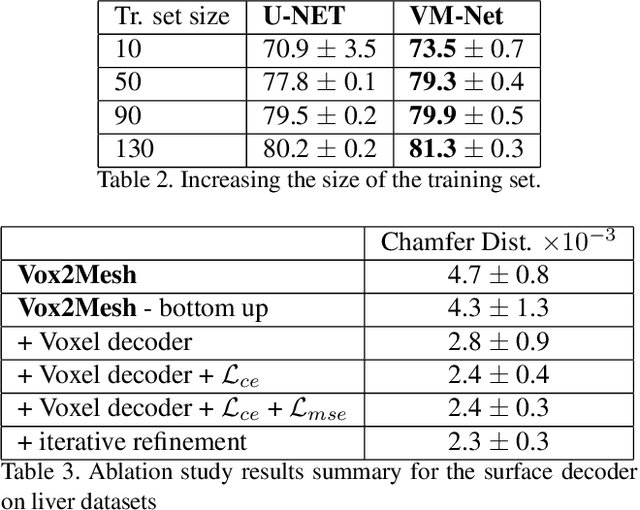
Abstract:CNN-based volumetric methods that label individual voxels now dominate the field of biomedical segmentation. In this paper, we show that simultaneously performing the segmentation and recovering a 3D mesh that models the surface can boost performance. To this end, we propose an end-to-end trainable two-stream encoder/decoder architecture. It comprises a single encoder and two decoders, one that labels voxels and the other outputs the mesh. The key to success is that the two decoders communicate with each other and help each other learn. This goes beyond the well-known fact that training a deep network to perform two different tasks improves its performance. We will demonstrate substantial performance increases on two very different and challenging datasets.
Probabilistic Atlases to Enforce Topological Constraints
Sep 18, 2019



Abstract:Probabilistic atlases (PAs) have long been used in standard segmentation approaches and, more recently, in conjunction with Convolutional Neural Networks (CNNs). However, their use has been restricted to relatively standardized structures such as the brain or heart which have limited or predictable range of deformations. Here we propose an encoding-decoding CNN architecture that can exploit rough atlases that encode only the topology of the target structures that can appear in any pose and have arbitrarily complex shapes to improve the segmentation results. It relies on the output of the encoder to compute both the pose parameters used to deform the atlas and the segmentation mask itself, which makes it effective and end-to-end trainable.
Modeling Brain Circuitry over a Wide Range of Scales
Apr 08, 2015



Abstract:If we are ever to unravel the mysteries of brain function at its most fundamental level, we will need a precise understanding of how its component neurons connect to each other. Electron Microscopes (EM) can now provide the nanometer resolution that is needed to image synapses, and therefore connections, while Light Microscopes (LM) see at the micrometer resolution required to model the 3D structure of the dendritic network. Since both the topology and the connection strength are integral parts of the brain's wiring diagram, being able to combine these two modalities is critically important. In fact, these microscopes now routinely produce high-resolution imagery in such large quantities that the bottleneck becomes automated processing and interpretation, which is needed for such data to be exploited to its full potential. In this paper, we briefly review the Computer Vision techniques we have developed at EPFL to address this need. They include delineating dendritic arbors from LM imagery, segmenting organelles from EM, and combining the two into a consistent representation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge