Giulio Guagliumi
Deep learning segmentation of fibrous cap in intravascular optical coherence tomography images
Nov 10, 2023Abstract:Thin-cap fibroatheroma (TCFA) is a prominent risk factor for plaque rupture. Intravascular optical coherence tomography (IVOCT) enables identification of fibrous cap (FC), measurement of FC thicknesses, and assessment of plaque vulnerability. We developed a fully-automated deep learning method for FC segmentation. This study included 32,531 images across 227 pullbacks from two registries. Images were semi-automatically labeled using our OCTOPUS with expert editing using established guidelines. We employed preprocessing including guidewire shadow detection, lumen segmentation, pixel-shifting, and Gaussian filtering on raw IVOCT (r,theta) images. Data were augmented in a natural way by changing theta in spiral acquisitions and by changing intensity and noise values. We used a modified SegResNet and comparison networks to segment FCs. We employed transfer learning from our existing much larger, fully-labeled calcification IVOCT dataset to reduce deep-learning training. Overall, our method consistently delivered better FC segmentation results (Dice: 0.837+/-0.012) than other deep-learning methods. Transfer learning reduced training time by 84% and reduced the need for more training samples. Our method showed a high level of generalizability, evidenced by highly-consistent segmentations across five-fold cross-validation (sensitivity: 85.0+/-0.3%, Dice: 0.846+/-0.011) and the held-out test (sensitivity: 84.9%, Dice: 0.816) sets. In addition, we found excellent agreement of FC thickness with ground truth (2.95+/-20.73 um), giving clinically insignificant bias. There was excellent reproducibility in pre- and post-stenting pullbacks (average FC angle: 200.9+/-128.0 deg / 202.0+/-121.1 deg). Our method will be useful for multiple research purposes and potentially for planning stent deployments that avoid placing a stent edge over an FC.
Automated segmentation of microvessels in intravascular OCT images using deep learning
Oct 01, 2022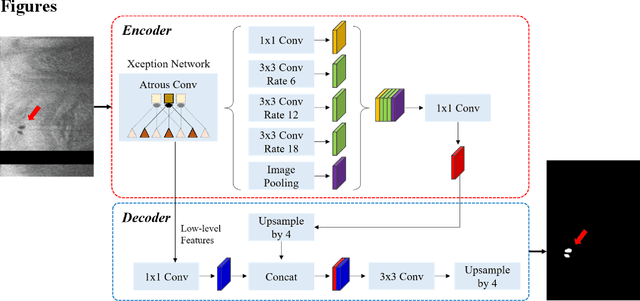
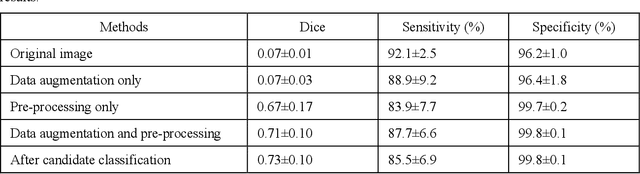

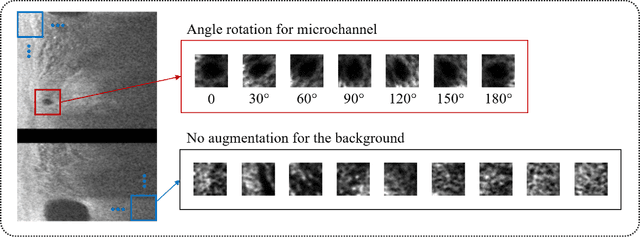
Abstract:To analyze this characteristic of vulnerability, we developed an automated deep learning method for detecting microvessels in intravascular optical coherence tomography (IVOCT) images. A total of 8,403 IVOCT image frames from 85 lesions and 37 normal segments were analyzed. Manual annotation was done using a dedicated software (OCTOPUS) previously developed by our group. Data augmentation in the polar (r,{\theta}) domain was applied to raw IVOCT images to ensure that microvessels appear at all possible angles. Pre-processing methods included guidewire/shadow detection, lumen segmentation, pixel shifting, and noise reduction. DeepLab v3+ was used to segment microvessel candidates. A bounding box on each candidate was classified as either microvessel or non-microvessel using a shallow convolutional neural network. For better classification, we used data augmentation (i.e., angle rotation) on bounding boxes with a microvessel during network training. Data augmentation and pre-processing steps improved microvessel segmentation performance significantly, yielding a method with Dice of 0.71+/-0.10 and pixel-wise sensitivity/specificity of 87.7+/-6.6%/99.8+/-0.1%. The network for classifying microvessels from candidates performed exceptionally well, with sensitivity of 99.5+/-0.3%, specificity of 98.8+/-1.0%, and accuracy of 99.1+/-0.5%. The classification step eliminated the majority of residual false positives, and the Dice coefficient increased from 0.71 to 0.73. In addition, our method produced 698 image frames with microvessels present, compared to 730 from manual analysis, representing a 4.4% difference. When compared to the manual method, the automated method improved microvessel continuity, implying improved segmentation performance. The method will be useful for research purposes as well as potential future treatment planning.
Automated analysis of fibrous cap in intravascular optical coherence tomography images of coronary arteries
Apr 21, 2022



Abstract:Thin-cap fibroatheroma (TCFA) and plaque rupture have been recognized as the most frequent risk factor for thrombosis and acute coronary syndrome. Intravascular optical coherence tomography (IVOCT) can identify TCFA and assess cap thickness, which provides an opportunity to assess plaque vulnerability. We developed an automated method that can detect lipidous plaque and assess fibrous cap thickness in IVOCT images. This study analyzed a total of 4,360 IVOCT image frames of 77 lesions among 41 patients. To improve segmentation performance, preprocessing included lumen segmentation, pixel-shifting, and noise filtering on the raw polar (r, theta) IVOCT images. We used the DeepLab-v3 plus deep learning model to classify lipidous plaque pixels. After lipid detection, we automatically detected the outer border of the fibrous cap using a special dynamic programming algorithm and assessed the cap thickness. Our method provided excellent discriminability of lipid plaque with a sensitivity of 85.8% and A-line Dice coefficient of 0.837. By comparing lipid angle measurements between two analysts following editing of our automated software, we found good agreement by Bland-Altman analysis (difference 6.7+/-17 degree; mean 196 degree). Our method accurately detected the fibrous cap from the detected lipid plaque. Automated analysis required a significant modification for only 5.5% frames. Furthermore, our method showed a good agreement of fibrous cap thickness between two analysts with Bland-Altman analysis (4.2+/-14.6 micron; mean 175 micron), indicating little bias between users and good reproducibility of the measurement. We developed a fully automated method for fibrous cap quantification in IVOCT images, resulting in good agreement with determinations by analysts. The method has great potential to enable highly automated, repeatable, and comprehensive evaluations of TCFAs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge