Gal Hyams
MicroAnalyzer: A Python Tool for Automated Bacterial Analysis with Fluorescence Microscopy
Sep 26, 2020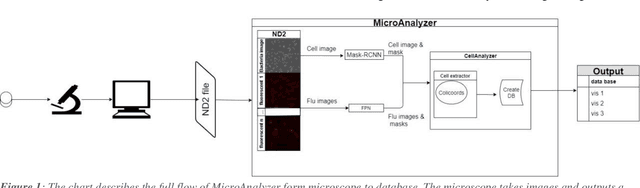
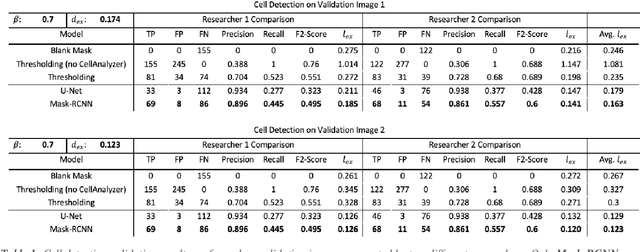
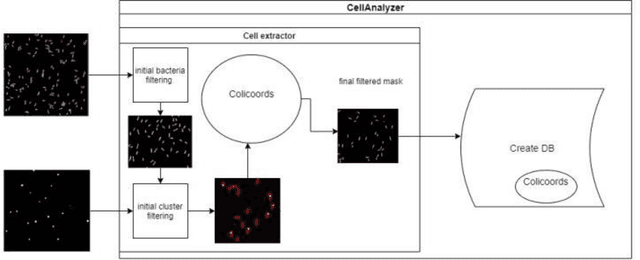
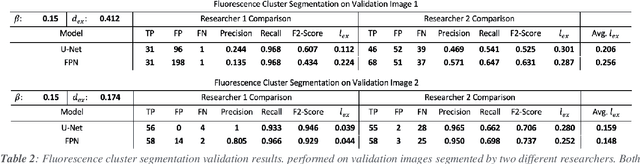
Abstract:Fluorescence microscopy is a widely used method among cell biologists for studying the localization and co-localization of fluorescent protein. For microbial cell biologists, these studies often include tedious and time-consuming manual segmentation of bacteria and of the fluorescence clusters or working with multiple programs. Here, we present MicroAnalyzer - a tool that automates these tasks by providing an end-to-end platform for microscope image analysis. While such tools do exist, they are costly, black-boxed programs. Microanalyzer offers an open-source alternative to these tools, allowing flexibility and expandability by advanced users. MicroAnalyzer provides accurate cell and fluorescence cluster segmentation based on state-of-the-art deep-learning segmentation models, combined with ad-hoc post-processing and Colicoords - an open-source cell image analysis tool for calculating general cell and fluorescence measurements. Using these methods, it performs better than generic approaches since the dynamic nature of neural networks allows for a quick adaptation to experiment restrictions and assumptions. Other existing tools do not consider experiment assumptions, nor do they provide fluorescence cluster detection without the need for any specialized equipment. The key goal of MicroAnalyzer is to automate the entire process of cell and fluorescence image analysis "from microscope to database", meaning it does not require any further input from the researcher except for the initial deep-learning model training. In this fashion, it allows the researchers to concentrate on the bigger picture instead of granular, eye-straining labor
Improved Training for Self-Training by Confidence Assessments
Apr 05, 2018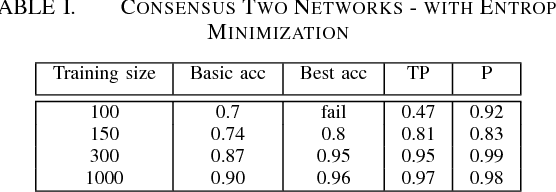
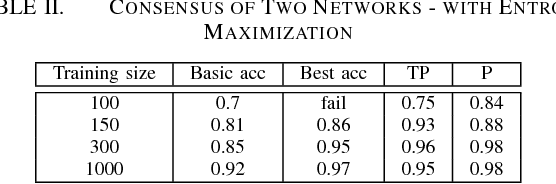


Abstract:It is well known that for some tasks, labeled data sets may be hard to gather. Therefore, we wished to tackle here the problem of having insufficient training data. We examined learning methods from unlabeled data after an initial training on a limited labeled data set. The suggested approach can be used as an online learning method on the unlabeled test set. In the general classification task, whenever we predict a label with high enough confidence, we treat it as a true label and train the data accordingly. For the semantic segmentation task, a classic example for an expensive data labeling process, we do so pixel-wise. Our suggested approaches were applied on the MNIST data-set as a proof of concept for a vision classification task and on the ADE20K data-set in order to tackle the semi-supervised semantic segmentation problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge