Florian Kowarsch
FATE: Feature-Agnostic Transformer-based Encoder for learning generalized embedding spaces in flow cytometry data
Nov 06, 2023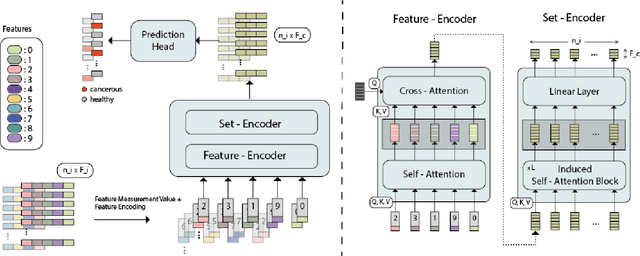
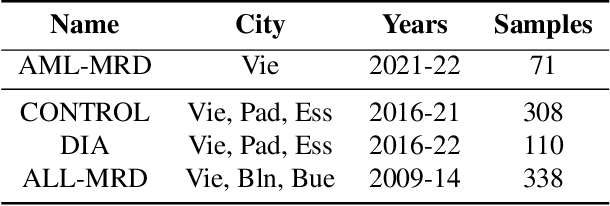
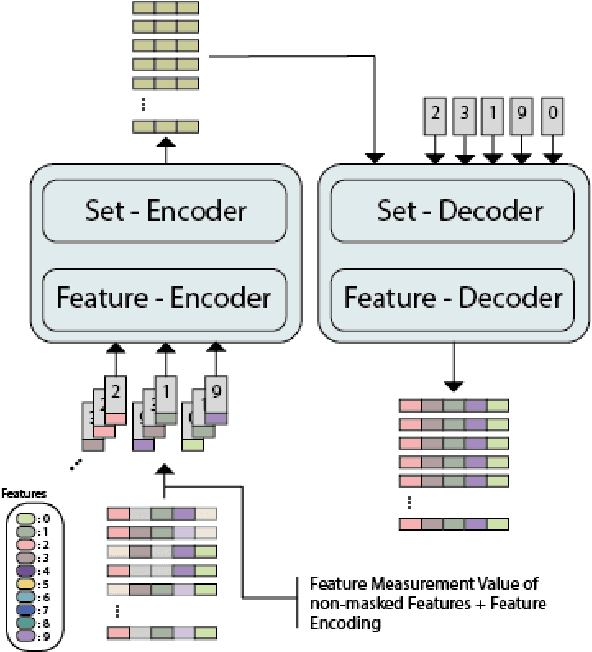
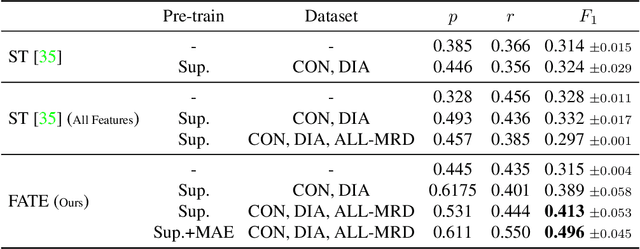
Abstract:While model architectures and training strategies have become more generic and flexible with respect to different data modalities over the past years, a persistent limitation lies in the assumption of fixed quantities and arrangements of input features. This limitation becomes particularly relevant in scenarios where the attributes captured during data acquisition vary across different samples. In this work, we aim at effectively leveraging data with varying features, without the need to constrain the input space to the intersection of potential feature sets or to expand it to their union. We propose a novel architecture that can directly process data without the necessity of aligned feature modalities by learning a general embedding space that captures the relationship between features across data samples with varying sets of features. This is achieved via a set-transformer architecture augmented by feature-encoder layers, thereby enabling the learning of a shared latent feature space from data originating from heterogeneous feature spaces. The advantages of the model are demonstrated for automatic cancer cell detection in acute myeloid leukemia in flow cytometry data, where the features measured during acquisition often vary between samples. Our proposed architecture's capacity to operate seamlessly across incongruent feature spaces is particularly relevant in this context, where data scarcity arises from the low prevalence of the disease. The code is available for research purposes at https://github.com/lisaweijler/FATE.
Explainable Techniques for Analyzing Flow Cytometry Cell Transformers
Jul 27, 2023



Abstract:Explainability for Deep Learning Models is especially important for clinical applications, where decisions of automated systems have far-reaching consequences. While various post-hoc explainable methods, such as attention visualization and saliency maps, already exist for common data modalities, including natural language and images, little work has been done to adapt them to the modality of Flow CytoMetry (FCM) data. In this work, we evaluate the usage of a transformer architecture called ReluFormer that ease attention visualization as well as we propose a gradient- and an attention-based visualization technique tailored for FCM. We qualitatively evaluate the visualization techniques for cell classification and polygon regression on pediatric Acute Lymphoblastic Leukemia (ALL) FCM samples. The results outline the model's decision process and demonstrate how to utilize the proposed techniques to inspect the trained model. The gradient-based visualization not only identifies cells that are most significant for a particular prediction but also indicates the directions in the FCM feature space in which changes have the most impact on the prediction. The attention visualization provides insights on the transformer's decision process when handling FCM data. We show that different attention heads specialize by attending to different biologically meaningful sub-populations in the data, even though the model retrieved solely supervised binary classification signals during training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge