Fanjin Wang
Filter Drug-induced Liver Injury Literature with Natural Language Processing and Ensemble Learning
Mar 09, 2022
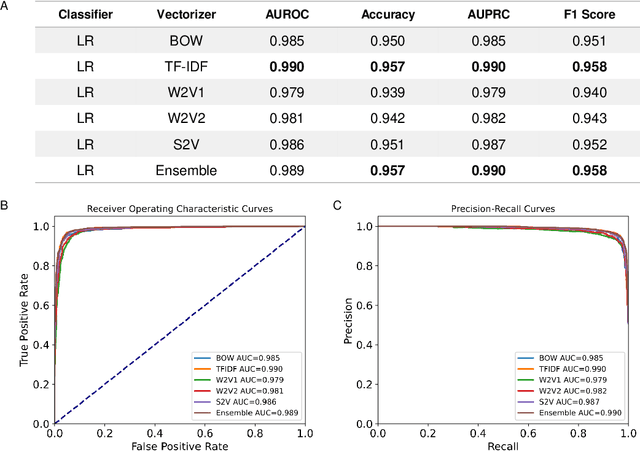
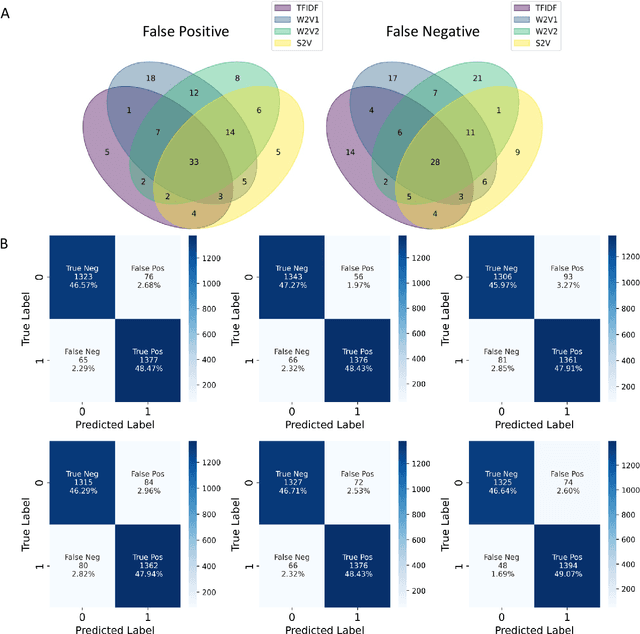
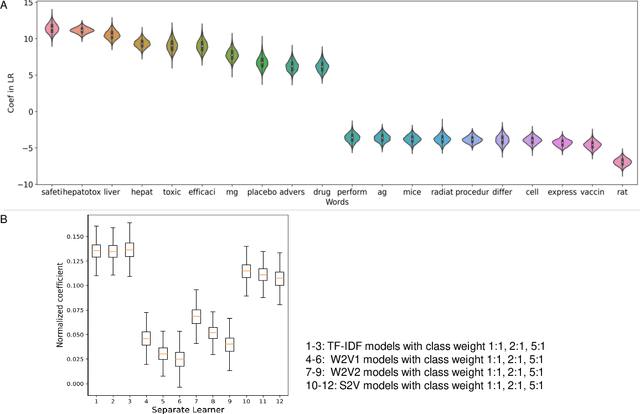
Abstract:Drug-induced liver injury (DILI) describes the adverse effects of drugs that damage liver. Life-threatening results including liver failure or death were also reported in severe DILI cases. Therefore, DILI-related events are strictly monitored for all approved drugs and the liver toxicity became important assessments for new drug candidates. These DILI-related reports are documented in hospital records, in clinical trial results, and also in research papers that contain preliminary in vitro and in vivo experiments. Conventionally, data extraction from previous publications relies heavily on resource-demanding manual labelling, which considerably decreased the efficiency of the information extraction process. The recent development of artificial intelligence, particularly, the rise of natural language processing (NLP) techniques, enabled the automatic processing of biomedical texts. In this study, based on around 28,000 papers (titles and abstracts) provided by the Critical Assessment of Massive Data Analysis (CAMDA) challenge, we benchmarked model performances on filtering out DILI literature. Among four word vectorization techniques, the model using term frequency-inverse document frequency (TF-IDF) and logistic regression outperformed others with an accuracy of 0.957 with our in-house test set. Furthermore, an ensemble model with similar overall performances was implemented and was fine-tuned to lower the false-negative cases to avoid neglecting potential DILI reports. The ensemble model achieved a high accuracy of 0.954 and an F1 score of 0.955 in the hold-out validation data provided by the CAMDA committee. Moreover, important words in positive/negative predictions were identified via model interpretation. Overall, the ensemble model reached satisfactory classification results, which can be further used by researchers to rapidly filter DILI-related literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge