Fabian Sinz
EMBC Special Issue: Calibrated Uncertainty for Trustworthy Clinical Gait Analysis Using Probabilistic Multiview Markerless Motion Capture
Jan 29, 2026Abstract:Video-based human movement analysis holds potential for movement assessment in clinical practice and research. However, the clinical implementation and trust of multi-view markerless motion capture (MMMC) require that, in addition to being accurate, these systems produce reliable confidence intervals to indicate how accurate they are for any individual. Building on our prior work utilizing variational inference to estimate joint angle posterior distributions, this study evaluates the calibration and reliability of a probabilistic MMMC method. We analyzed data from 68 participants across two institutions, validating the model against an instrumented walkway and standard marker-based motion capture. We measured the calibration of the confidence intervals using the Expected Calibration Error (ECE). The model demonstrated reliable calibration, yielding ECE values generally < 0.1 for both step and stride length and bias-corrected gait kinematics. We observed a median step and stride length error of ~16 mm and ~12 mm respectively, with median bias-corrected kinematic errors ranging from 1.5 to 3.8 degrees across lower extremity joints. Consistent with the calibrated ECE, the magnitude of the model's predicted uncertainty correlated strongly with observed error measures. These findings indicate that, as designed, the probabilistic model reconstruction quantifies epistemic uncertainty, allowing it to identify unreliable outputs without the need for concurrent ground-truth instrumentation.
Biomechanical Reconstruction with Confidence Intervals from Multiview Markerless Motion Capture
Feb 10, 2025



Abstract:Advances in multiview markerless motion capture (MMMC) promise high-quality movement analysis for clinical practice and research. While prior validation studies show MMMC performs well on average, they do not provide what is needed in clinical practice or for large-scale utilization of MMMC -- confidence intervals over specific kinematic estimates from a specific individual analyzed using a possibly unique camera configuration. We extend our previous work using an implicit representation of trajectories optimized end-to-end through a differentiable biomechanical model to learn the posterior probability distribution over pose given all the detected keypoints. This posterior probability is learned through a variational approximation and estimates confidence intervals for individual joints at each moment in a trial, showing confidence intervals generally within 10-15 mm of spatial error for virtual marker locations, consistent with our prior validation studies. Confidence intervals over joint angles are typically only a few degrees and widen for more distal joints. The posterior also models the correlation structure over joint angles, such as correlations between hip and pelvis angles. The confidence intervals estimated through this method allow us to identify times and trials where kinematic uncertainty is high.
Adversarial Distribution Balancing for Counterfactual Reasoning
Nov 28, 2023



Abstract:The development of causal prediction models is challenged by the fact that the outcome is only observable for the applied (factual) intervention and not for its alternatives (the so-called counterfactuals); in medicine we only know patients' survival for the administered drug and not for other therapeutic options. Machine learning approaches for counterfactual reasoning have to deal with both unobserved outcomes and distributional differences due to non-random treatment administration. Unsupervised domain adaptation (UDA) addresses similar issues; one has to deal with unobserved outcomes -- the labels of the target domain -- and distributional differences between source and target domain. We propose Adversarial Distribution Balancing for Counterfactual Reasoning (ADBCR), which directly uses potential outcome estimates of the counterfactuals to remove spurious causal relations. We show that ADBCR outcompetes state-of-the-art methods on three benchmark datasets, and demonstrate that ADBCR's performance can be further improved if unlabeled validation data are included in the training procedure to better adapt the model to the validation domain.
In All Likelihood, Deep Belief Is Not Enough
Nov 28, 2010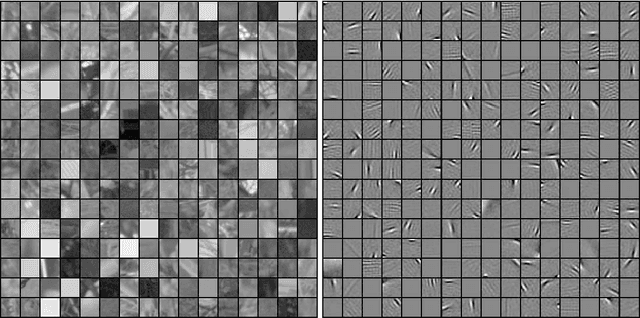
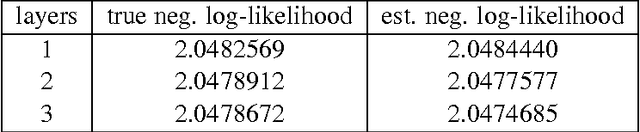
Abstract:Statistical models of natural stimuli provide an important tool for researchers in the fields of machine learning and computational neuroscience. A canonical way to quantitatively assess and compare the performance of statistical models is given by the likelihood. One class of statistical models which has recently gained increasing popularity and has been applied to a variety of complex data are deep belief networks. Analyses of these models, however, have been typically limited to qualitative analyses based on samples due to the computationally intractable nature of the model likelihood. Motivated by these circumstances, the present article provides a consistent estimator for the likelihood that is both computationally tractable and simple to apply in practice. Using this estimator, a deep belief network which has been suggested for the modeling of natural image patches is quantitatively investigated and compared to other models of natural image patches. Contrary to earlier claims based on qualitative results, the results presented in this article provide evidence that the model under investigation is not a particularly good model for natural images
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge