Eric Jul
Deep Learning and Computer Vision Techniques for Microcirculation Analysis: A Review
May 11, 2022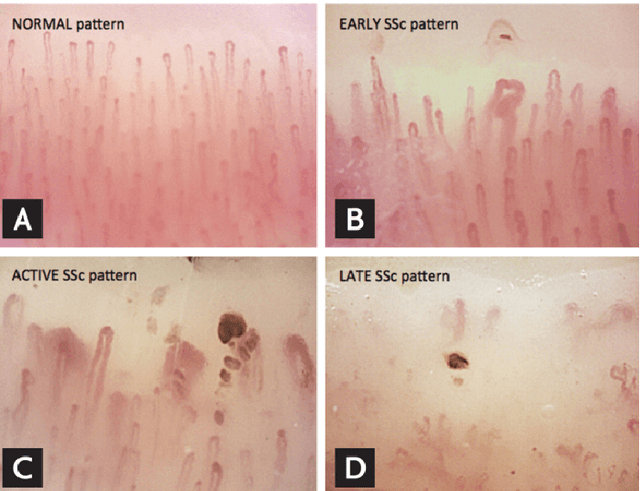

Abstract:The analysis of microcirculation images has the potential to reveal early signs of life-threatening diseases like sepsis. Quantifying the capillary density and the capillary distribution in microcirculation images can be used as a biological marker to assist critically ill patients. The quantification of these biological markers is labor-intensive, time-consuming, and subject to interobserver variability. Several computer vision techniques with varying performance can be used to automate the analysis of these microcirculation images in light of the stated challenges. In this paper, we present a survey of over 50 research papers and present the most relevant and promising computer vision algorithms to automate the analysis of microcirculation images. Furthermore, we present a survey of the methods currently used by other researchers to automate the analysis of microcirculation images. This survey is of high clinical relevance because it acts as a guidebook of techniques for other researchers to develop their microcirculation analysis systems and algorithms.
CapillaryX: A Software Design Pattern for Analyzing Medical Images in Real-time using Deep Learning
Apr 13, 2022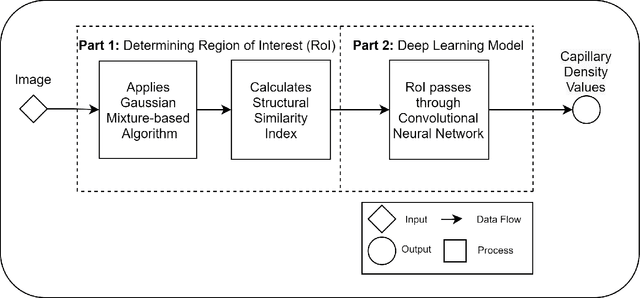
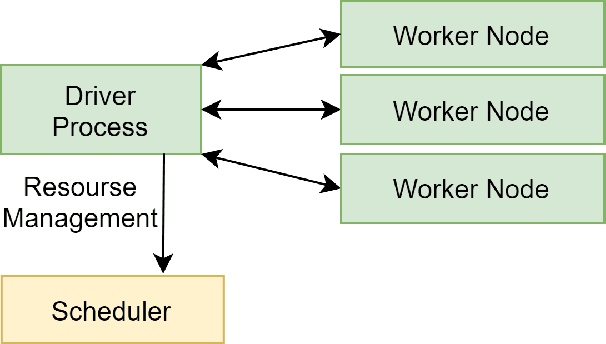
Abstract:Recent advances in digital imaging, e.g., increased number of pixels captured, have meant that the volume of data to be processed and analyzed from these images has also increased. Deep learning algorithms are state-of-the-art for analyzing such images, given their high accuracy when trained with a large data volume of data. Nevertheless, such analysis requires considerable computational power, making such algorithms time- and resource-demanding. Such high demands can be met by using third-party cloud service providers. However, analyzing medical images using such services raises several legal and privacy challenges and does not necessarily provide real-time results. This paper provides a computing architecture that locally and in parallel can analyze medical images in real-time using deep learning thus avoiding the legal and privacy challenges stemming from uploading data to a third-party cloud provider. To make local image processing efficient on modern multi-core processors, we utilize parallel execution to offset the resource-intensive demands of deep neural networks. We focus on a specific medical-industrial case study, namely the quantifying of blood vessels in microcirculation images for which we have developed a working system. It is currently used in an industrial, clinical research setting as part of an e-health application. Our results show that our system is approximately 78% faster than its serial system counterpart and 12% faster than a master-slave parallel system architecture.
CapillaryNet: An Automated System to Analyze Microcirculation Videos from Handheld Vital Microscopy
May 21, 2021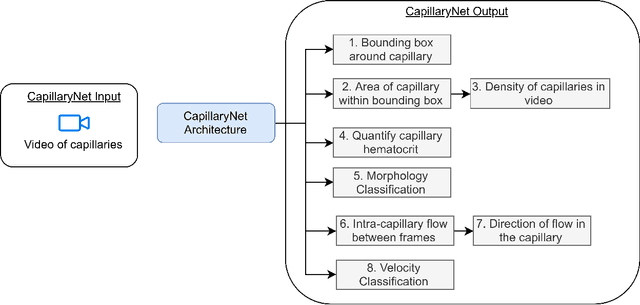

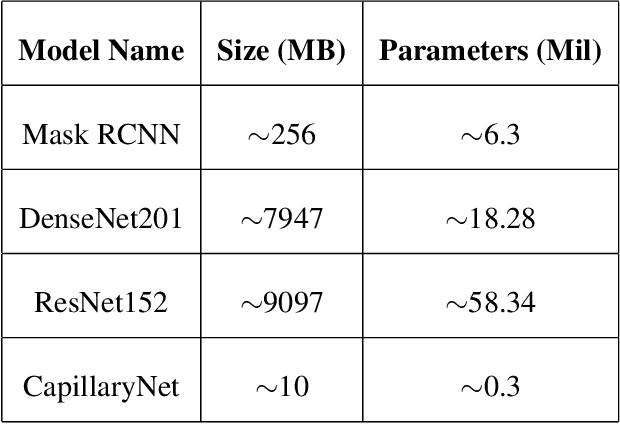
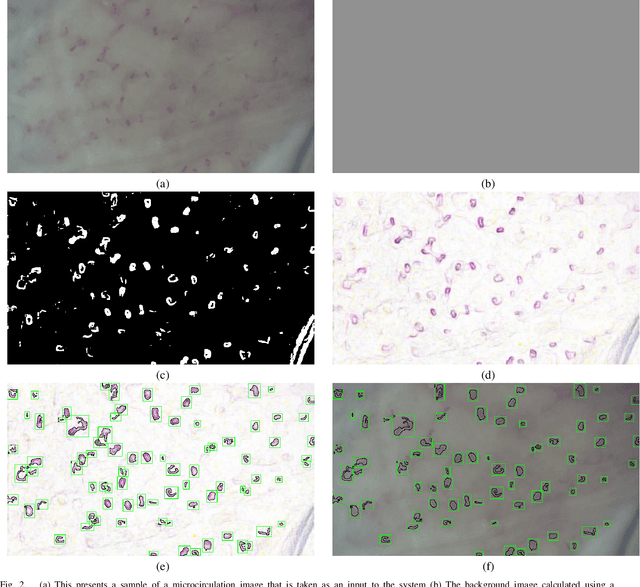
Abstract:Capillaries are the smallest vessels in the body responsible for the delivery of oxygen and nutrients to the surrounding cells. Various diseases have been shown to alter the density of nutritive capillaries and the flow velocity of erythrocytes. In previous studies, capillary density and flow velocity have been assessed manually by trained specialists. Manual analysis of a 20-second long microvascular video takes on average 20 minutes and requires extensive training. Several studies have reported that manual analysis hinders the application of microvascular microscopy in a clinical setting. In this paper, we present a fully automated system, called CapillaryNet, that can automate microvascular microscopy analysis so it can be used as a clinical application. Moreover, CapillaryNet measures several microvascular parameters that researchers were previously unable to quantify, i.e. capillary hematocrit and intra-capillary flow velocity heterogeneity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge