Ekaterina Gracheva
Light and Accurate: Neural Architecture Search via Two Constant Shared Weights Initialisations
Feb 09, 2023Abstract:In recent years, zero-cost proxies are gaining ground in neural architecture search (NAS). These methods allow finding the optimal neural network for a given task faster and with a lesser computational load than conventional NAS methods. Equally important is the fact that they also shed some light on the internal workings of neural architectures. This paper presents a zero-cost metric that highly correlates with the train set accuracy across the NAS-Bench-101, NAS-Bench-201 and NAS-Bench-NLP benchmark datasets. Architectures are initialised with two distinct constant shared weights, one at a time. Then, a fixed random mini-batch of data is passed forward through each initialisation. We observe that the dispersion of the outputs between two initialisations positively correlates with trained accuracy. The correlation further improves when we normalise dispersion by average output magnitude. Our metric, epsilon, does not require gradients computation or labels. It thus unbinds the NAS procedure from training hyperparameters, loss metrics and human-labelled data. Our method is easy to integrate within existing NAS algorithms and takes a fraction of a second to evaluate a single network.
Trainless Model Performance Estimation for Neural Architecture Search
Mar 10, 2021



Abstract:Neural architecture search has become an indispensable part of the deep learning field. Modern methods allow to find out the best performing architectures for a task, or to build a network from scratch, but they usually require a tremendous amount of training. In this paper we present a simple method, allowing to discover a suitable architecture for a task based on its untrained performance. We introduce the metric score as the relative standard deviation of the untrained accuracy, which is the standard deviation divided by the mean. Statistics for each neural architecture are calculated over multiple initialisations with different seeds on a single batch of data. An architecture with the lowest metric score value has on average an accuracy of $91.90 \pm 2.27$, $64.08 \pm 5.63$ and $38.76 \pm 6.62$ for CIFAR-10, CIFAR-100 and a downscaled version of ImageNet, respectively. The results show that a good architecture should be stable against initialisations before training. The procedure takes about $190$ s for CIFAR and $133.9$ s for ImageNet, on a batch of $256$ images and $100$ initialisations.
SMILES-X: autonomous molecular compounds characterization for small datasets without descriptors
Jul 04, 2019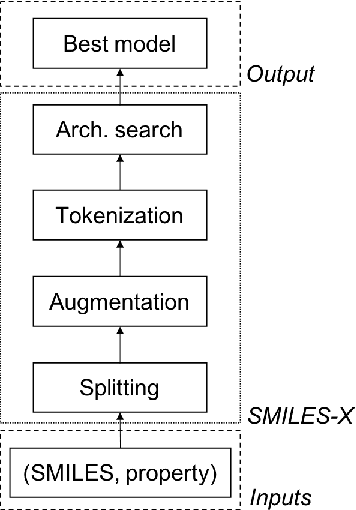

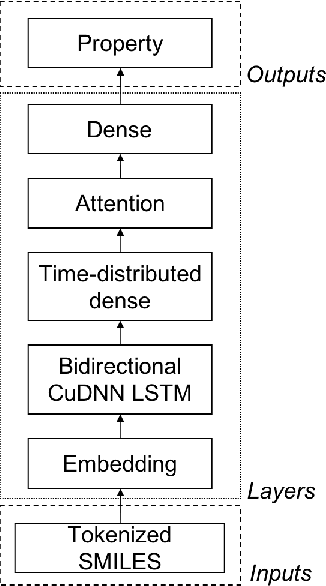
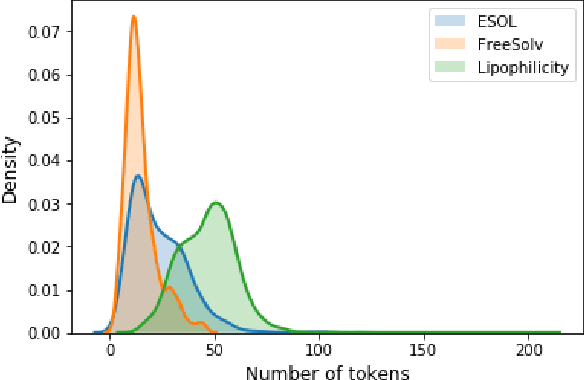
Abstract:There is more and more evidence that machine learning can be successfully applied in materials science and related fields. However, datasets in these fields are often quite small ($\ll1000$ samples). It makes the most advanced machine learning techniques remain neglected, as they are considered to be applicable to big data only. Moreover, materials informatics methods often rely on human-engineered descriptors, that should be carefully chosen, or even created, to fit the physicochemical property that one intends to predict. In this article, we propose a new method that tackles both the issue of small datasets and the difficulty of task-specific descriptors development. The SMILES-X is an autonomous pipeline for molecular compounds characterisation based on a \{Embed-Encode-Attend-Predict\} neural architecture with a data-specific Bayesian hyper-parameters optimisation. The only input to the architecture -- the SMILES strings -- are de-canonicalised in order to efficiently augment the data. One of the key features of the architecture is the attention mechanism, which enables the interpretation of output predictions without extra computational cost. The SMILES-X shows new state-of-the-art results in the inference of aqueous solubility ($\overline{RMSE}_{test} \simeq 0.57 \pm 0.07$ mols/L), hydration free energy ($\overline{RMSE}_{test} \simeq 0.81 \pm 0.22$ kcal/mol, which is $\sim 24.5\%$ better than molecular dynamics simulations), and octanol/water distribution coefficient ($\overline{RMSE}_{test} \simeq 0.59 \pm 0.02$ for LogD at pH 7.4) of molecular compounds. The SMILES-X is intended to become an important asset in the toolkit of materials scientists and chemists. The source code for the SMILES-X is available at \href{https://github.com/GLambard/SMILES-X}{github.com/GLambard/SMILES-X}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge