Edyta Szurowska
BRONCO: Automated modelling of the bronchovascular bundle using the Computed Tomography Images
Sep 18, 2023
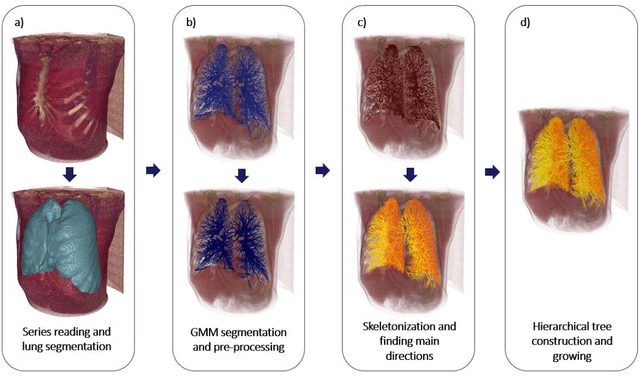
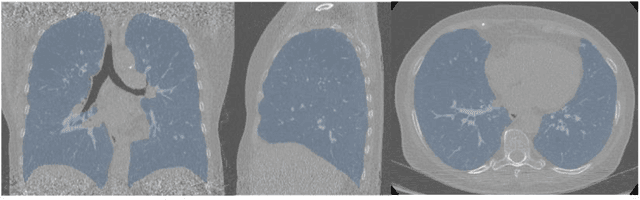
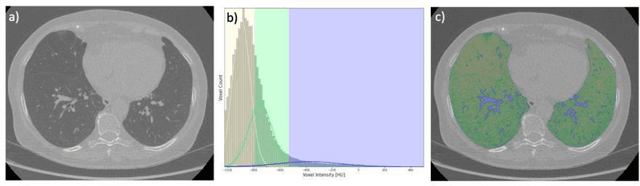
Abstract:Segmentation of the bronchovascular bundle within the lung parenchyma is a key step for the proper analysis and planning of many pulmonary diseases. It might also be considered the preprocessing step when the goal is to segment the nodules from the lung parenchyma. We propose a segmentation pipeline for the bronchovascular bundle based on the Computed Tomography images, returning either binary or labelled masks of vessels and bronchi situated in the lung parenchyma. The method consists of two modules, modeling of the bronchial tree and vessels. The core revolves around a similar pipeline, the determination of the initial perimeter by the GMM method, skeletonization, and hierarchical analysis of the created graph. We tested our method on both low-dose CT and standard-dose CT, with various pathologies, reconstructed with various slice thicknesses, and acquired from various machines. We conclude that the method is invariant with respect to the origin and parameters of the CT series. Our pipeline is best suited for studies with healthy patients, patients with lung nodules, and patients with emphysema.
Review of methods for automatic cerebral microbleeds detection
Jan 31, 2023Abstract:Cerebral microbleeds detection is an important and challenging task. With the gaining popularity of the MRI, the ability to detect cerebral microbleeds also raises. Unfortunately, for radiologists, it is a time-consuming and laborious procedure. For this reason, various solutions to automate this process have been proposed for several years, but none of them is currently used in medical practice. In this context, the need to systematize the existing knowledge and best practices has been recognized as a factor facilitating the imminent synthesis of a real CMBs detection system practically applicable in medicine. To the best of our knowledge, all available publications regarding automatic cerebral microbleeds detection have been gathered, described, and assessed in this paper in order to distinguish the current research state and provide a starting point for future studies.
POLCOVID: a multicenter multiclass chest X-ray database
Dec 15, 2022Abstract:The outbreak of the SARS-CoV-2 pandemic has put healthcare systems worldwide to their limits, resulting in increased waiting time for diagnosis and required medical assistance. With chest radiographs (CXR) being one of the most common COVID-19 diagnosis methods, many artificial intelligence tools for image-based COVID-19 detection have been developed, often trained on a small number of images from COVID-19-positive patients. Thus, the need for high-quality and well-annotated CXR image databases increased. This paper introduces POLCOVID dataset, containing chest X-ray (CXR) images of patients with COVID-19 or other-type pneumonia, and healthy individuals gathered from 15 Polish hospitals. The original radiographs are accompanied by the preprocessed images limited to the lung area and the corresponding lung masks obtained with the segmentation model. Moreover, the manually created lung masks are provided for a part of POLCOVID dataset and the other four publicly available CXR image collections. POLCOVID dataset can help in pneumonia or COVID-19 diagnosis, while the set of matched images and lung masks may serve for the development of lung segmentation solutions.
CIRCA: comprehensible online system in support of chest X-rays-based COVID-19 diagnosis
Oct 11, 2022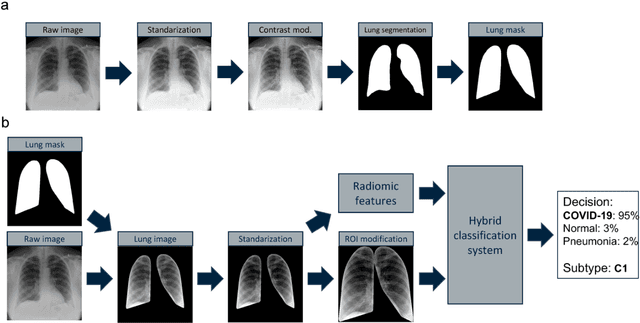
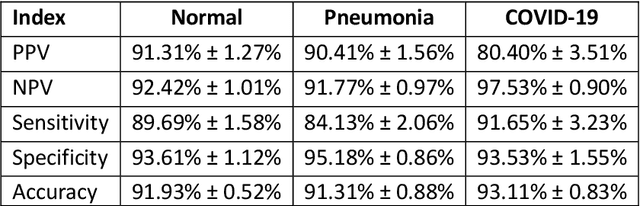
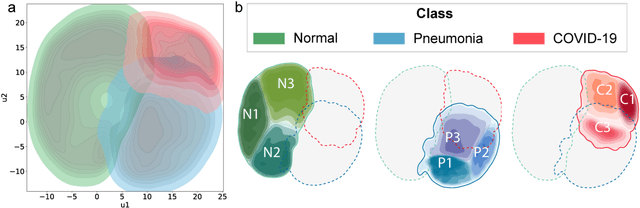
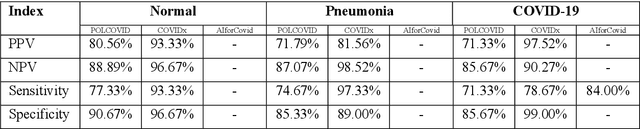
Abstract:Due to the large accumulation of patients requiring hospitalization, the COVID-19 pandemic disease caused a high overload of health systems, even in developed countries. Deep learning techniques based on medical imaging data can help in the faster detection of COVID-19 cases and monitoring of disease progression. Regardless of the numerous proposed solutions for lung X-rays, none of them is a product that can be used in the clinic. Five different datasets (POLCOVID, AIforCOVID, COVIDx, NIH, and artificially generated data) were used to construct a representative dataset of 23 799 CXRs for model training; 1 050 images were used as a hold-out test set, and 44 247 as independent test set (BIMCV database). A U-Net-based model was developed to identify a clinically relevant region of the CXR. Each image class (normal, pneumonia, and COVID-19) was divided into 3 subtypes using a 2D Gaussian mixture model. A decision tree was used to aggregate predictions from the InceptionV3 network based on processed CXRs and a dense neural network on radiomic features. The lung segmentation model gave the Sorensen-Dice coefficient of 94.86% in the validation dataset, and 93.36% in the testing dataset. In 5-fold cross-validation, the accuracy for all classes ranged from 91% to 93%, keeping slightly higher specificity than sensitivity and NPV than PPV. In the hold-out test set, the balanced accuracy ranged between 68% and 100%. The highest performance was obtained for the subtypes N1, P1, and C1. A similar performance was obtained on the independent dataset for normal and COVID-19 class subtypes. Seventy-six percent of COVID-19 patients wrongly classified as normal cases were annotated by radiologists as with no signs of disease. Finally, we developed the online service (https://circa.aei.polsl.pl) to provide access to fast diagnosis support tools.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge