Denis Reilly
Detection of Obstructive Sleep Apnoea Using Features Extracted from Segmented Time-Series ECG Signals Using a One Dimensional Convolutional Neural Network
Feb 03, 2020



Abstract:The study in this paper presents a one-dimensional convolutional neural network (1DCNN) model, designed for the automated detection of obstructive Sleep Apnoea (OSA) captured from single-channel electrocardiogram (ECG) signals. The system provides mechanisms in clinical practice that help diagnose patients suffering with OSA. Using the state-of-the-art in 1DCNNs, a model is constructed using convolutional, max pooling layers and a fully connected Multilayer Perceptron (MLP) consisting of a hidden layer and SoftMax output for classification. The 1DCNN extracts prominent features, which are used to train an MLP. The model is trained using segmented ECG signals grouped into 5 unique datasets of set window sizes. 35 ECG signal recordings were selected from an annotated database containing 70 night-time ECG recordings. (Group A = a01 to a20 (Apnoea breathing), Group B = b01 to b05 (moderate), and Group C = c01 to c10 (normal). A total of 6514 minutes of Apnoea was recorded. Evaluation of the model is performed using a set of standard metrics which show the proposed model achieves high classification results in both training and validation using our windowing strategy, particularly W=500 (Sensitivity 0.9705, Specificity 0.9725, F1 Score 0.9717, Kappa Score 0.9430, Log Loss 0.0836, ROCAUC 0.9945). This demonstrates the model can identify the presence of Apnoea with a high degree of accuracy.
SAERMA: Stacked Autoencoder Rule Mining Algorithm for the Interpretation of Epistatic Interactions in GWAS for Extreme Obesity
Aug 27, 2019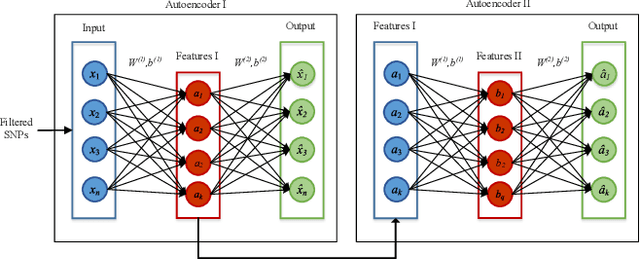
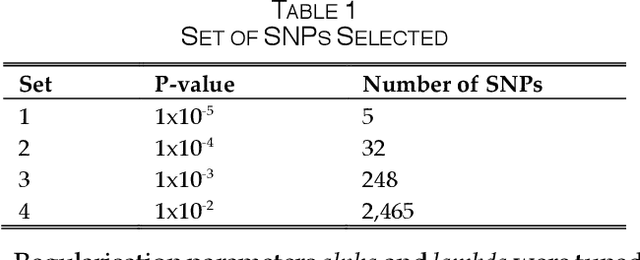
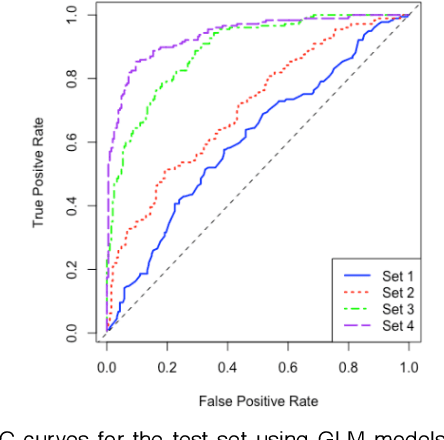
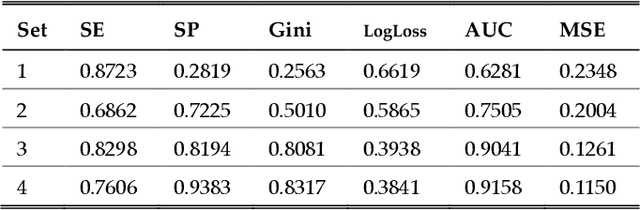
Abstract:One of the most important challenges in the analysis of high-throughput genetic data is the development of efficient computational methods to identify statistically significant Single Nucleotide Polymorphisms (SNPs). Genome-wide association studies (GWAS) use single-locus analysis where each SNP is independently tested for association with phenotypes. The limitation with this approach, however, is its inability to explain genetic variation in complex diseases. Alternative approaches are required to model the intricate relationships between SNPs. Our proposed approach extends GWAS by combining deep learning stacked autoencoders (SAEs) and association rule mining (ARM) to identify epistatic interactions between SNPs. Following traditional GWAS quality control and association analysis, the most significant SNPs are selected and used in the subsequent analysis to investigate epistasis. SAERMA controls the classification results produced in the final fully connected multi-layer feedforward artificial neural network (MLP) by manipulating the interestingness measures, support and confidence, in the rule generation process. The best classification results were achieved with 204 SNPs compressed to 100 units (77% AUC, 77% SE, 68% SP, 53% Gini, logloss=0.58, and MSE=0.20), although it was possible to achieve 73% AUC (77% SE, 63% SP, 45% Gini, logloss=0.62, and MSE=0.21) with 50 hidden units - both supported by close model interpretation.
Modelling Segmented Cardiotocography Time-Series Signals Using One-Dimensional Convolutional Neural Networks for the Early Detection of Abnormal Birth Outcomes
Aug 06, 2019



Abstract:Gynaecologists and obstetricians visually interpret cardiotocography (CTG) traces using the International Federation of Gynaecology and Obstetrics (FIGO) guidelines to assess the wellbeing of the foetus during antenatal care. This approach has raised concerns among professionals concerning inter- and intra-variability where clinical diagnosis only has a 30% positive predictive value when classifying pathological outcomes. Machine learning models, trained with FIGO and other user derived features extracted from CTG traces, have been shown to increase positive predictive capacity and minimise variability. This is only possible however when class distributions are equal which is rarely the case in clinical trials where case-control observations are heavily skewed. Classes can be balanced using either synthetic data derived from resampled case training data or by decreasing the number of control instances. However, this introduces bias and removes valuable information. Concerns have also been raised regarding machine learning studies and their reliance on manually handcrafted features. While this has led to some interesting results, deriving an optimal set of features is considered to be an art as well as a science and is often an empirical and time consuming process. In this paper, we address both of these issues and propose a novel CTG analysis methodology that a) splits CTG time series signals into n-size windows with equal class distributions, and b) automatically extracts features from time-series windows using a one dimensional convolutional neural network (1DCNN) and multilayer perceptron (MLP) ensemble. Our proposed method achieved good results using a window size of 200 with (Sens=0.7981, Spec=0.7881, F1=0.7830, Kappa=0.5849, AUC=0.8599, and Logloss=0.4791).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge