David M. Rocke
GPU-Accelerated Primal Learning for Extremely Fast Large-Scale Classification
Aug 08, 2020

Abstract:One of the most efficient methods to solve L2-regularized primal problems, such as logistic regression and linear support vector machine (SVM) classification, is the widely used trust region Newton algorithm, TRON. While TRON has recently been shown to enjoy substantial speedups on shared-memory multi-core systems, exploiting graphical processing units (GPUs) to speed up the method is significantly more difficult, owing to the highly complex and heavily sequential nature of the algorithm. In this work, we show that using judicious GPU-optimization principles, TRON training time for different losses and feature representations may be drastically reduced. For sparse feature sets, we show that using GPUs to train logistic regression classifiers in LIBLINEAR is up to an order-of-magnitude faster than solely using multithreading. For dense feature sets--which impose far more stringent memory constraints--we show that GPUs substantially reduce the lengthy SVM learning times required for state-of-the-art proteomics analysis, leading to dramatic improvements over recently proposed speedups. Furthermore, we show how GPU speedups may be mixed with multithreading to enable such speedups when the dataset is too large for GPU memory requirements; on a massive dense proteomics dataset of nearly a quarter-billion data instances, these mixed-architecture speedups reduce SVM analysis time from over half a week to less than a single day while using limited GPU memory.
Learning Concave Conditional Likelihood Models for Improved Analysis of Tandem Mass Spectra
Sep 04, 2019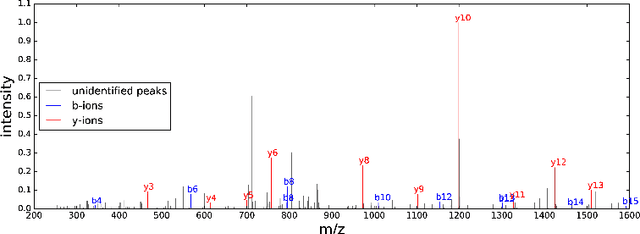
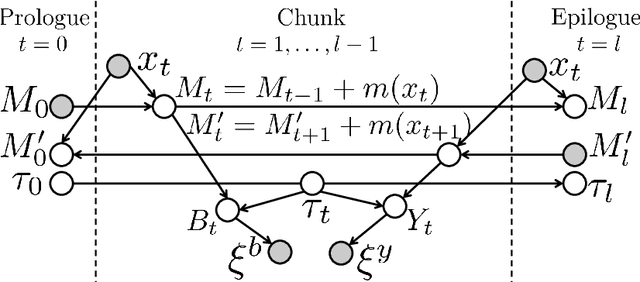
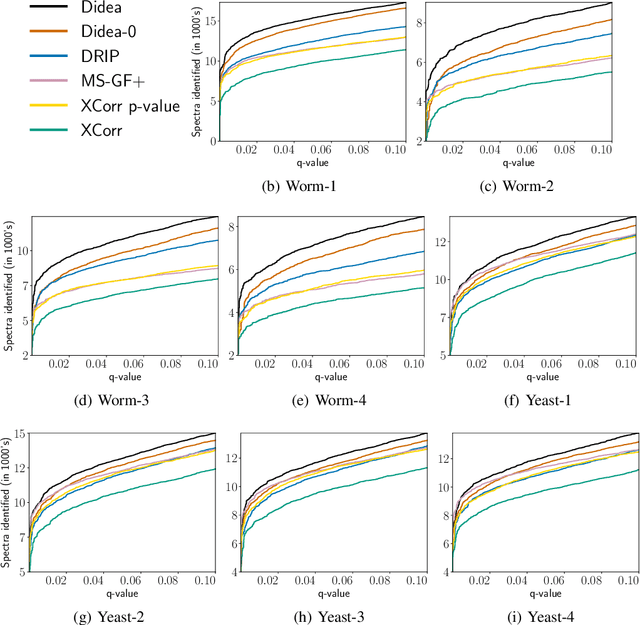
Abstract:The most widely used technology to identify the proteins present in a complex biological sample is tandem mass spectrometry, which quickly produces a large collection of spectra representative of the peptides (i.e., protein subsequences) present in the original sample. In this work, we greatly expand the parameter learning capabilities of a dynamic Bayesian network (DBN) peptide-scoring algorithm, Didea, by deriving emission distributions for which its conditional log-likelihood scoring function remains concave. We show that this class of emission distributions, called Convex Virtual Emissions (CVEs), naturally generalizes the log-sum-exp function while rendering both maximum likelihood estimation and conditional maximum likelihood estimation concave for a wide range of Bayesian networks. Utilizing CVEs in Didea allows efficient learning of a large number of parameters while ensuring global convergence, in stark contrast to Didea's previous parameter learning framework (which could only learn a single parameter using a costly grid search) and other trainable models (which only ensure convergence to local optima). The newly trained scoring function substantially outperforms the state-of-the-art in both scoring function accuracy and downstream Fisher kernel analysis. Furthermore, we significantly improve Didea's runtime performance through successive optimizations to its message passing schedule and derive explicit connections between Didea's new concave score and related MS/MS scoring functions.
Gradients of Generative Models for Improved Discriminative Analysis of Tandem Mass Spectra
Sep 04, 2019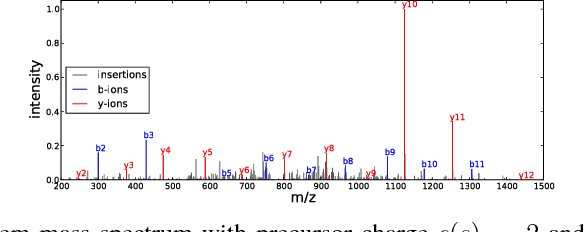
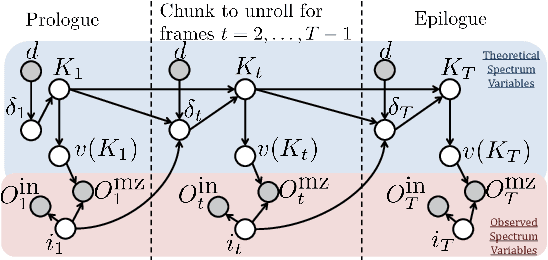
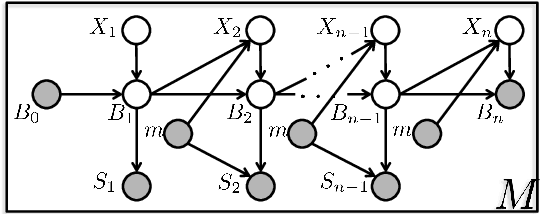
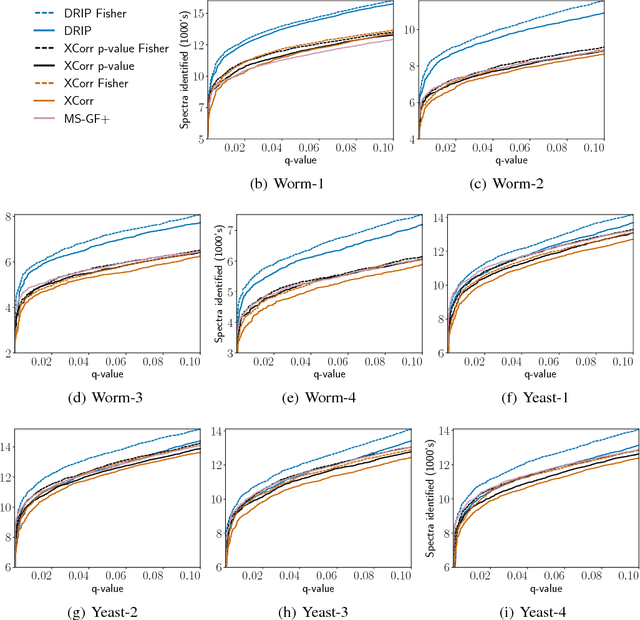
Abstract:Tandem mass spectrometry (MS/MS) is a high-throughput technology used toidentify the proteins in a complex biological sample, such as a drop of blood. A collection of spectra is generated at the output of the process, each spectrum of which is representative of a peptide (protein subsequence) present in the original complex sample. In this work, we leverage the log-likelihood gradients of generative models to improve the identification of such spectra. In particular, we show that the gradient of a recently proposed dynamic Bayesian network (DBN) may be naturally employed by a kernel-based discriminative classifier. The resulting Fisher kernel substantially improves upon recent attempts to combine generative and discriminative models for post-processing analysis, outperforming all other methods on the evaluated datasets. We extend the improved accuracy offered by the Fisher kernel framework to other search algorithms by introducing Theseus, a DBN representing a large number of widely used MS/MS scoring functions. Furthermore, with gradient ascent and max-product inference at hand, we use Theseus to learn model parameters without any supervision.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge