Gradients of Generative Models for Improved Discriminative Analysis of Tandem Mass Spectra
Paper and Code
Sep 04, 2019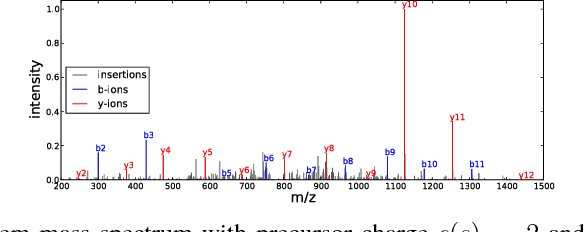
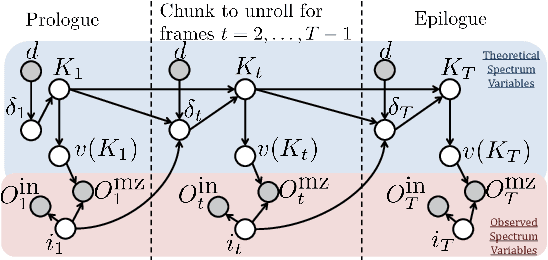
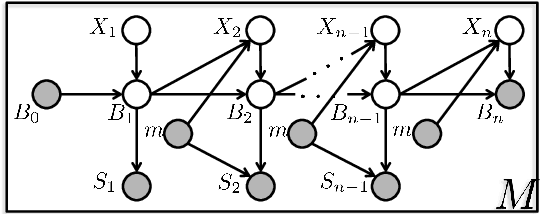
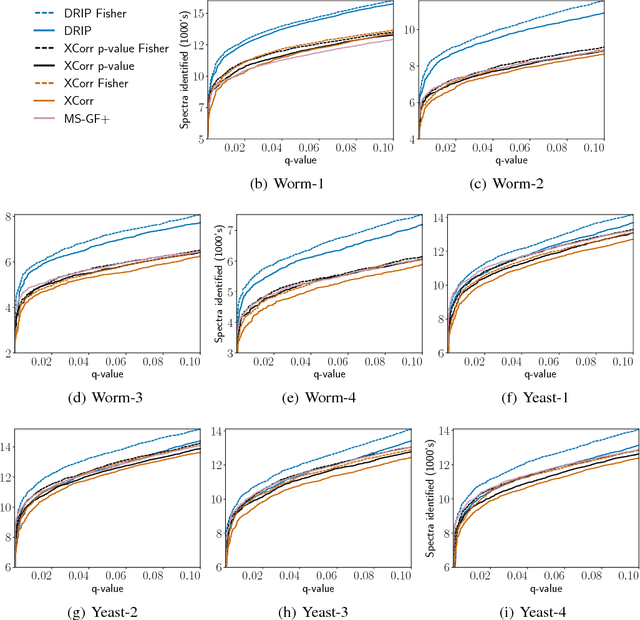
Tandem mass spectrometry (MS/MS) is a high-throughput technology used toidentify the proteins in a complex biological sample, such as a drop of blood. A collection of spectra is generated at the output of the process, each spectrum of which is representative of a peptide (protein subsequence) present in the original complex sample. In this work, we leverage the log-likelihood gradients of generative models to improve the identification of such spectra. In particular, we show that the gradient of a recently proposed dynamic Bayesian network (DBN) may be naturally employed by a kernel-based discriminative classifier. The resulting Fisher kernel substantially improves upon recent attempts to combine generative and discriminative models for post-processing analysis, outperforming all other methods on the evaluated datasets. We extend the improved accuracy offered by the Fisher kernel framework to other search algorithms by introducing Theseus, a DBN representing a large number of widely used MS/MS scoring functions. Furthermore, with gradient ascent and max-product inference at hand, we use Theseus to learn model parameters without any supervision.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge