David Buterez
Masked Attention is All You Need for Graphs
Feb 16, 2024Abstract:Graph neural networks (GNNs) and variations of the message passing algorithm are the predominant means for learning on graphs, largely due to their flexibility, speed, and satisfactory performance. The design of powerful and general purpose GNNs, however, requires significant research efforts and often relies on handcrafted, carefully-chosen message passing operators. Motivated by this, we propose a remarkably simple alternative for learning on graphs that relies exclusively on attention. Graphs are represented as node or edge sets and their connectivity is enforced by masking the attention weight matrix, effectively creating custom attention patterns for each graph. Despite its simplicity, masked attention for graphs (MAG) has state-of-the-art performance on long-range tasks and outperforms strong message passing baselines and much more involved attention-based methods on over 55 node and graph-level tasks. We also show significantly better transfer learning capabilities compared to GNNs and comparable or better time and memory scaling. MAG has sub-linear memory scaling in the number of nodes or edges, enabling learning on dense graphs and future-proofing the approach.
Graph Neural Networks with Adaptive Readouts
Nov 09, 2022Abstract:An effective aggregation of node features into a graph-level representation via readout functions is an essential step in numerous learning tasks involving graph neural networks. Typically, readouts are simple and non-adaptive functions designed such that the resulting hypothesis space is permutation invariant. Prior work on deep sets indicates that such readouts might require complex node embeddings that can be difficult to learn via standard neighborhood aggregation schemes. Motivated by this, we investigate the potential of adaptive readouts given by neural networks that do not necessarily give rise to permutation invariant hypothesis spaces. We argue that in some problems such as binding affinity prediction where molecules are typically presented in a canonical form it might be possible to relax the constraints on permutation invariance of the hypothesis space and learn a more effective model of the affinity by employing an adaptive readout function. Our empirical results demonstrate the effectiveness of neural readouts on more than 40 datasets spanning different domains and graph characteristics. Moreover, we observe a consistent improvement over standard readouts (i.e., sum, max, and mean) relative to the number of neighborhood aggregation iterations and different convolutional operators.
Efficient approximation of DNA hybridisation using deep learning
Feb 19, 2021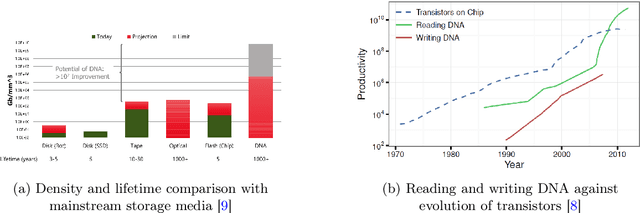
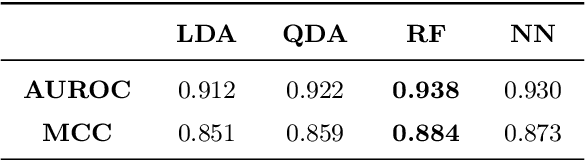
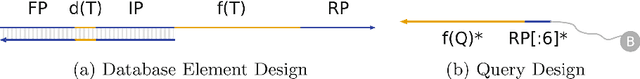
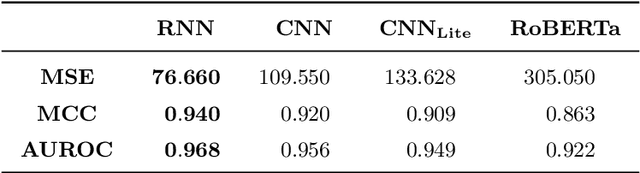
Abstract:Deoxyribonucleic acid (DNA) has shown great promise in enabling computational applications, most notably in the fields of DNA data storage and DNA computing. The former exploits the natural properties of DNA, such as high storage density and longevity, for the archival of digital information, while the latter aims to use the interactivity of DNA to encode computations. Recently, the two paradigms were jointly used to formulate the near-data processing concept for DNA databases, where the computations are performed directly on the stored data. The fundamental, low-level operation that DNA naturally possesses is that of hybridisation, also called annealing, of complementary sequences. Information is encoded as DNA strands, which will naturally bind in solution, thus enabling search and pattern-matching capabilities. Being able to control and predict the process of hybridisation is crucial for the ambitious future of the so-called Hybrid Molecular-Electronic Computing. Current tools are, however, limited in terms of throughput and applicability to large-scale problems. In this work, we present the first comprehensive study of machine learning methods applied to the task of predicting DNA hybridisation. For this purpose, we introduce a synthetic hybridisation dataset of over 2.5 million data points, enabling the use of a wide range of machine learning algorithms, including the latest in deep learning. Depending on the hardware, the proposed models provide a reduction in inference time ranging from one to over two orders of magnitude compared to the state-of-the-art, while retaining high fidelity. We then discuss the integration of our methods in modern, scalable workflows. The implementation is available at: https://github.com/davidbuterez/dna-hyb-deep-learning
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge