Darren Treanor
National Pathology Imaging Cooperative, Leeds Teaching Hospitals NHS Trust, Leeds, UK, University of Leeds, Leeds, UK, Linköping University, Linköping, Sweden
Machine learning-based multimodal prognostic models integrating pathology images and high-throughput omic data for overall survival prediction in cancer: a systematic review
Jul 22, 2025Abstract:Multimodal machine learning integrating histopathology and molecular data shows promise for cancer prognostication. We systematically reviewed studies combining whole slide images (WSIs) and high-throughput omics to predict overall survival. Searches of EMBASE, PubMed, and Cochrane CENTRAL (12/08/2024), plus citation screening, identified eligible studies. Data extraction used CHARMS; bias was assessed with PROBAST+AI; synthesis followed SWiM and PRISMA 2020. Protocol: PROSPERO (CRD42024594745). Forty-eight studies (all since 2017) across 19 cancer types met criteria; all used The Cancer Genome Atlas. Approaches included regularised Cox regression (n=4), classical ML (n=13), and deep learning (n=31). Reported c-indices ranged 0.550-0.857; multimodal models typically outperformed unimodal ones. However, all studies showed unclear/high bias, limited external validation, and little focus on clinical utility. Multimodal WSI-omics survival prediction is a fast-growing field with promising results but needs improved methodological rigor, broader datasets, and clinical evaluation. Funded by NPIC, Leeds Teaching Hospitals NHS Trust, UK (Project 104687), supported by UKRI Industrial Strategy Challenge Fund.
Artificial intelligence in digital pathology: a diagnostic test accuracy systematic review and meta-analysis
Jun 19, 2023Abstract:Ensuring diagnostic performance of AI models before clinical use is key to the safe and successful adoption of these technologies. Studies reporting AI applied to digital pathology images for diagnostic purposes have rapidly increased in number in recent years. The aim of this work is to provide an overview of the diagnostic accuracy of AI in digital pathology images from all areas of pathology. This systematic review and meta-analysis included diagnostic accuracy studies using any type of artificial intelligence applied to whole slide images (WSIs) in any disease type. The reference standard was diagnosis through histopathological assessment and / or immunohistochemistry. Searches were conducted in PubMed, EMBASE and CENTRAL in June 2022. We identified 2976 studies, of which 100 were included in the review and 48 in the full meta-analysis. Risk of bias and concerns of applicability were assessed using the QUADAS-2 tool. Data extraction was conducted by two investigators and meta-analysis was performed using a bivariate random effects model. 100 studies were identified for inclusion, equating to over 152,000 whole slide images (WSIs) and representing many disease types. Of these, 48 studies were included in the meta-analysis. These studies reported a mean sensitivity of 96.3% (CI 94.1-97.7) and mean specificity of 93.3% (CI 90.5-95.4) for AI. There was substantial heterogeneity in study design and all 100 studies identified for inclusion had at least one area at high or unclear risk of bias. This review provides a broad overview of AI performance across applications in whole slide imaging. However, there is huge variability in study design and available performance data, with details around the conduct of the study and make up of the datasets frequently missing. Overall, AI offers good accuracy when applied to WSIs but requires more rigorous evaluation of its performance.
Navigating the reporting guideline environment for computational pathology: A review
Jan 03, 2023Abstract:The application of new artificial intelligence (AI) discoveries is transforming healthcare research. However, the standards of reporting are variable in this still evolving field, leading to potential research waste. The aim of this work is to highlight resources and reporting guidelines available to researchers working in computational pathology. The EQUATOR Network library of reporting guidelines and extensions was systematically searched up to August 2022 to identify applicable resources. Inclusion and exclusion criteria were used and guidance was screened for utility at different stages of research and for a range of study types. Items were compiled to create a summary for easy identification of useful resources and guidance. Over 70 published resources applicable to pathology AI research were identified. Guidelines were divided into key categories, reflecting current study types and target areas for AI research: Literature & Research Priorities, Discovery, Clinical Trial, Implementation and Post-Implementation & Guidelines. Guidelines useful at multiple stages of research and those currently in development were also highlighted. Summary tables with links to guidelines for these groups were developed, to assist those working in cancer AI research with complete reporting of research. Issues with replication and research waste are recognised problems in AI research. Reporting guidelines can be used as templates to ensure the essential information needed to replicate research is included within journal articles and abstracts. Reporting guidelines are available and useful for many study types, but greater awareness is needed to encourage researchers to utilise them and for journals to adopt them. This review and summary of resources highlights guidance to researchers, aiming to improve completeness of reporting.
Weakly-supervised learning for image-based classification of primary melanomas into genomic immune subgroups
Feb 23, 2022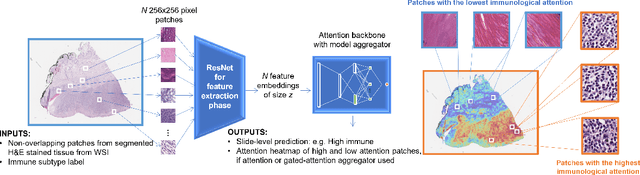

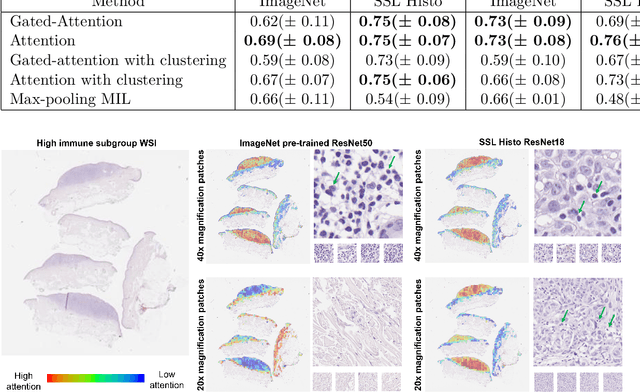
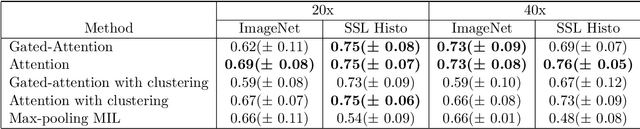
Abstract:Determining early-stage prognostic markers and stratifying patients for effective treatment are two key challenges for improving outcomes for melanoma patients. Previous studies have used tumour transcriptome data to stratify patients into immune subgroups, which were associated with differential melanoma specific survival and potential treatment strategies. However, acquiring transcriptome data is a time-consuming and costly process. Moreover, it is not routinely used in the current clinical workflow. Here we attempt to overcome this by developing deep learning models to classify gigapixel H&E stained pathology slides, which are well established in clinical workflows, into these immune subgroups. Previous subtyping approaches have employed supervised learning which requires fully annotated data, or have only examined single genetic mutations in melanoma patients. We leverage a multiple-instance learning approach, which only requires slide-level labels and uses an attention mechanism to highlight regions of high importance to the classification. Moreover, we show that pathology-specific self-supervised models generate better representations compared to pathology-agnostic models for improving our model performance, achieving a mean AUC of 0.76 for classifying histopathology images as high or low immune subgroups. We anticipate that this method may allow us to find new biomarkers of high importance and could act as a tool for clinicians to infer the immune landscape of tumours and stratify patients, without needing to carry out additional expensive genetic tests.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge