Daniel Hidru
Dr.VAE: Drug Response Variational Autoencoder
Jul 06, 2017



Abstract:We present two deep generative models based on Variational Autoencoders to improve the accuracy of drug response prediction. Our models, Perturbation Variational Autoencoder and its semi-supervised extension, Drug Response Variational Autoencoder (Dr.VAE), learn latent representation of the underlying gene states before and after drug application that depend on: (i) drug-induced biological change of each gene and (ii) overall treatment response outcome. Our VAE-based models outperform the current published benchmarks in the field by anywhere from 3 to 11% AUROC and 2 to 30% AUPR. In addition, we found that better reconstruction accuracy does not necessarily lead to improvement in classification accuracy and that jointly trained models perform better than models that minimize reconstruction error independently.
EquiNMF: Graph Regularized Multiview Nonnegative Matrix Factorization
Sep 14, 2014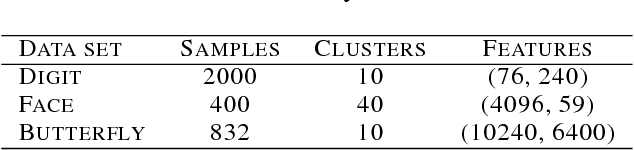
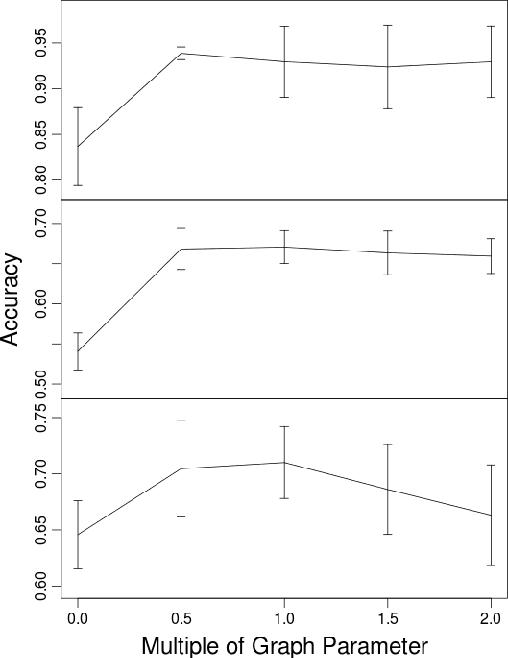
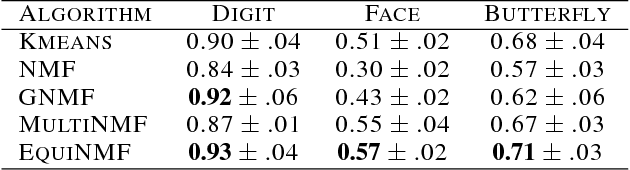
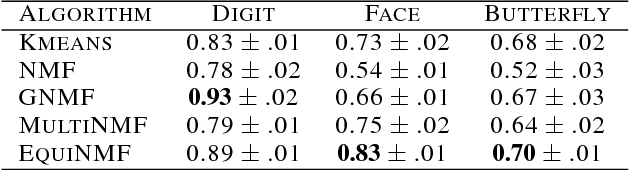
Abstract:Nonnegative matrix factorization (NMF) methods have proved to be powerful across a wide range of real-world clustering applications. Integrating multiple types of measurements for the same objects/subjects allows us to gain a deeper understanding of the data and refine the clustering. We have developed a novel Graph-reguarized multiview NMF-based method for data integration called EquiNMF. The parameters for our method are set in a completely automated data-specific unsupervised fashion, a highly desirable property in real-world applications. We performed extensive and comprehensive experiments on multiview imaging data. We show that EquiNMF consistently outperforms other single-view NMF methods used on concatenated data and multi-view NMF methods with different types of regularizations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge