Damayanthi Herath
Semantic Segmentation using Vision Transformers: A survey
May 05, 2023Abstract:Semantic segmentation has a broad range of applications in a variety of domains including land coverage analysis, autonomous driving, and medical image analysis. Convolutional neural networks (CNN) and Vision Transformers (ViTs) provide the architecture models for semantic segmentation. Even though ViTs have proven success in image classification, they cannot be directly applied to dense prediction tasks such as image segmentation and object detection since ViT is not a general purpose backbone due to its patch partitioning scheme. In this survey, we discuss some of the different ViT architectures that can be used for semantic segmentation and how their evolution managed the above-stated challenge. The rise of ViT and its performance with a high success rate motivated the community to slowly replace the traditional convolutional neural networks in various computer vision tasks. This survey aims to review and compare the performances of ViT architectures designed for semantic segmentation using benchmarking datasets. This will be worthwhile for the community to yield knowledge regarding the implementations carried out in semantic segmentation and to discover more efficient methodologies using ViTs.
Machine learning for plant microRNA prediction: A systematic review
Jun 29, 2021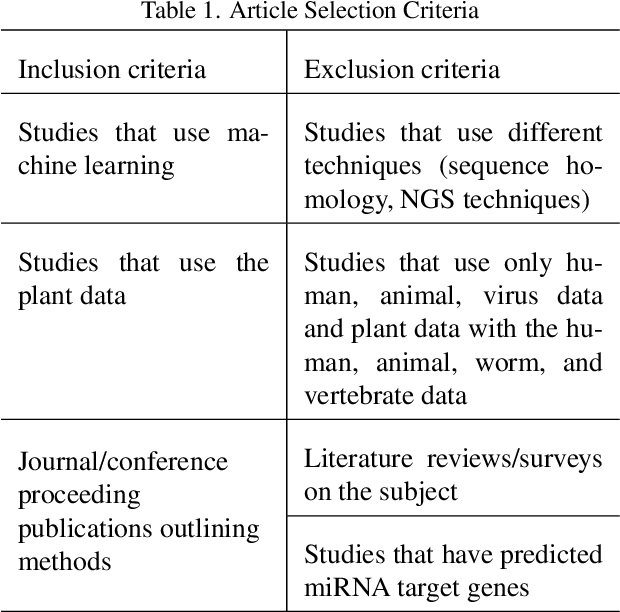
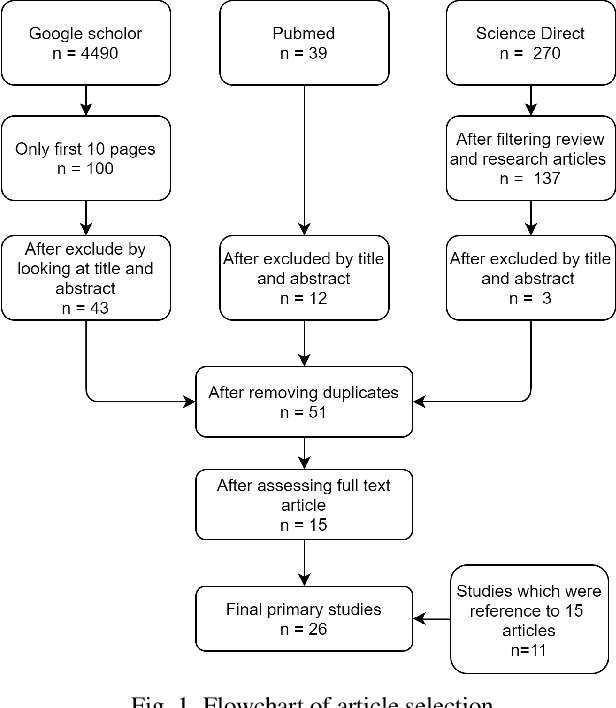
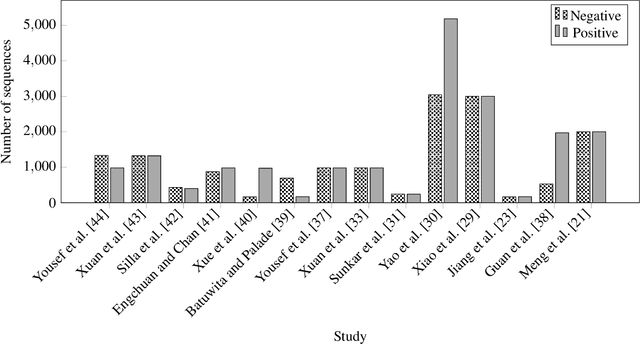
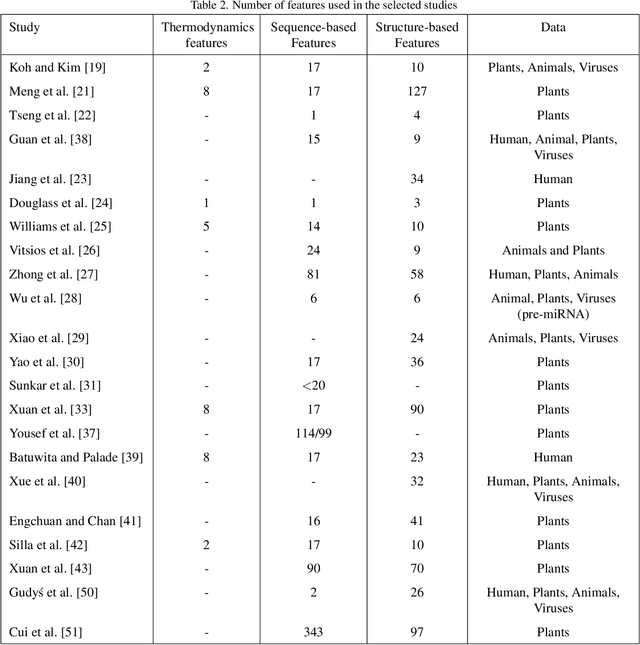
Abstract:MicroRNAs (miRNAs) are endogenous small non-coding RNAs that play an important role in post-transcriptional gene regulation. However, the experimental determination of miRNA sequence and structure is both expensive and time-consuming. Therefore, computational and machine learning-based approaches have been adopted to predict novel microRNAs. With the involvement of data science and machine learning in biology, multiple research studies have been conducted to find microRNAs with different computational methods and different miRNA features. Multiple approaches are discussed in detail considering the learning algorithm/s used, features considered, dataset/s used and the criteria used in evaluations. This systematic review focuses on the machine learning methods developed for miRNA identification in plants. This will help researchers to gain a detailed idea about past studies and identify novel paths that solve drawbacks occurred in past studies. Our findings highlight the need for plant-specific computational methods for miRNA identification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge