Dániel Unyi
Neurodevelopmental Phenotype Prediction: A State-of-the-Art Deep Learning Model
Nov 16, 2022Abstract:A major challenge in medical image analysis is the automated detection of biomarkers from neuroimaging data. Traditional approaches, often based on image registration, are limited in capturing the high variability of cortical organisation across individuals. Deep learning methods have been shown to be successful in overcoming this difficulty, and some of them have even outperformed medical professionals on certain datasets. In this paper, we apply a deep neural network to analyse the cortical surface data of neonates, derived from the publicly available Developing Human Connectome Project (dHCP). Our goal is to identify neurodevelopmental biomarkers and to predict gestational age at birth based on these biomarkers. Using scans of preterm neonates acquired around the term-equivalent age, we were able to investigate the impact of preterm birth on cortical growth and maturation during late gestation. Besides reaching state-of-the-art prediction accuracy, the proposed model has much fewer parameters than the baselines, and its error stays low on both unregistered and registered cortical surfaces.
Utility of Equivariant Message Passing in Cortical Mesh Segmentation
Jun 15, 2022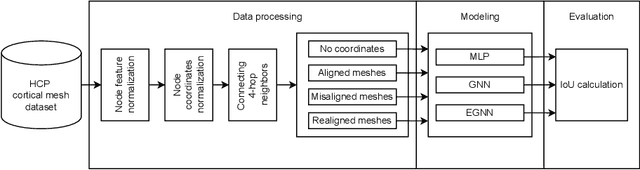
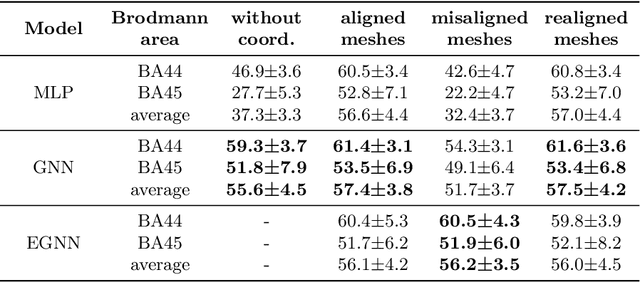
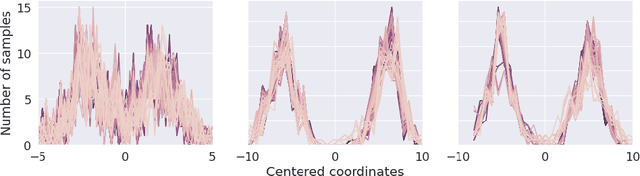
Abstract:The automated segmentation of cortical areas has been a long-standing challenge in medical image analysis. The complex geometry of the cortex is commonly represented as a polygon mesh, whose segmentation can be addressed by graph-based learning methods. When cortical meshes are misaligned across subjects, current methods produce significantly worse segmentation results, limiting their ability to handle multi-domain data. In this paper, we investigate the utility of E(n)-equivariant graph neural networks (EGNNs), comparing their performance against plain graph neural networks (GNNs). Our evaluation shows that GNNs outperform EGNNs on aligned meshes, due to their ability to leverage the presence of a global coordinate system. On misaligned meshes, the performance of plain GNNs drop considerably, while E(n)-equivariant message passing maintains the same segmentation results. The best results can also be obtained by using plain GNNs on realigned data (co-registered meshes in a global coordinate system).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge