Connor Robertson
Advancing calibration for stochastic agent-based models in epidemiology with Stein variational inference and Gaussian process surrogates
Feb 26, 2025
Abstract:Accurate calibration of stochastic agent-based models (ABMs) in epidemiology is crucial to make them useful in public health policy decisions and interventions. Traditional calibration methods, e.g., Markov Chain Monte Carlo (MCMC), that yield a probability density function for the parameters being calibrated, are often computationally expensive. When applied to ABMs which are highly parametrized, the calibration process becomes computationally infeasible. This paper investigates the utility of Stein Variational Inference (SVI) as an alternative calibration technique for stochastic epidemiological ABMs approximated by Gaussian process (GP) surrogates. SVI leverages gradient information to iteratively update a set of particles in the space of parameters being calibrated, offering potential advantages in scalability and efficiency for high-dimensional ABMs. The ensemble of particles yields a joint probability density function for the parameters and serves as the calibration. We compare the performance of SVI and MCMC in calibrating CityCOVID, a stochastic epidemiological ABM, focusing on predictive accuracy and calibration effectiveness. Our results demonstrate that SVI maintains predictive accuracy and calibration effectiveness comparable to MCMC, making it a viable alternative for complex epidemiological models. We also present the practical challenges of using a gradient-based calibration such as SVI which include careful tuning of hyperparameters and monitoring of the particle dynamics.
Bayesian calibration of stochastic agent based model via random forest
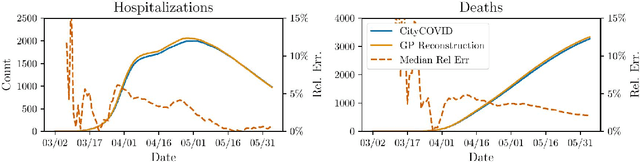
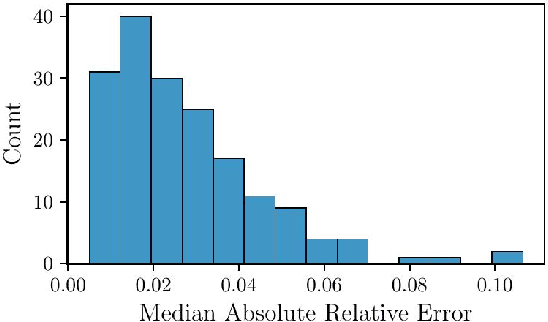
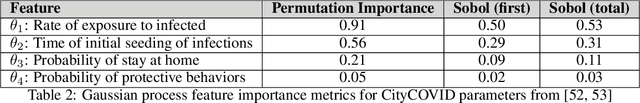
Jun 27, 2024Abstract:Agent-based models (ABM) provide an excellent framework for modeling outbreaks and interventions in epidemiology by explicitly accounting for diverse individual interactions and environments. However, these models are usually stochastic and highly parametrized, requiring precise calibration for predictive performance. When considering realistic numbers of agents and properly accounting for stochasticity, this high dimensional calibration can be computationally prohibitive. This paper presents a random forest based surrogate modeling technique to accelerate the evaluation of ABMs and demonstrates its use to calibrate an epidemiological ABM named CityCOVID via Markov chain Monte Carlo (MCMC). The technique is first outlined in the context of CityCOVID's quantities of interest, namely hospitalizations and deaths, by exploring dimensionality reduction via temporal decomposition with principal component analysis (PCA) and via sensitivity analysis. The calibration problem is then presented and samples are generated to best match COVID-19 hospitalization and death numbers in Chicago from March to June in 2020. These results are compared with previous approximate Bayesian calibration (IMABC) results and their predictive performance is analyzed showing improved performance with a reduction in computation.
Performing Video Frame Prediction of Microbial Growth with a Recurrent Neural Network
May 12, 2022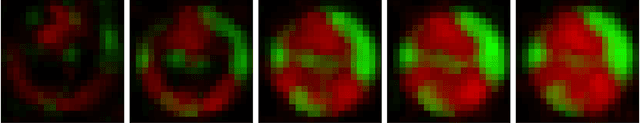
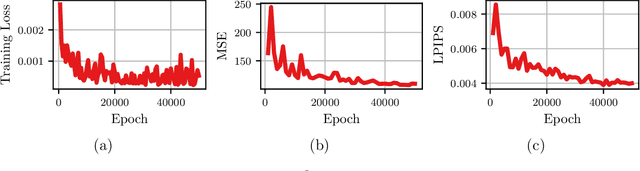
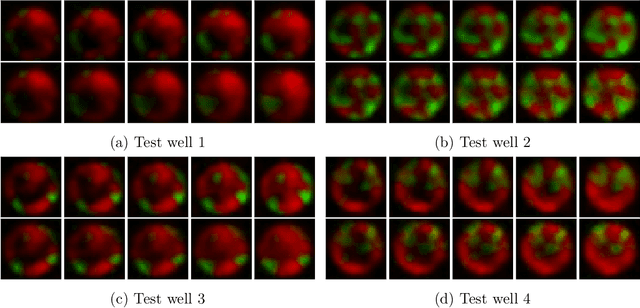
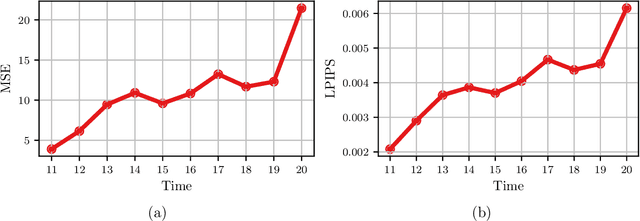
Abstract:A Recurrent Neural Network (RNN) was used to perform video frame prediction of microbial growth for a population of two mutants of Pseudomonas aeruginosa. The RNN was trained on videos of 20 frames that were acquired using fluorescence microscopy and microfluidics. The network predicted the last 10 frames of each video, and the accuracy's of the predictions was assessed by comparing raw images, population curves, and the number and size of individual colonies. Overall, we found the predictions to be accurate using this approach. The implications this result has on designing autonomous experiments in microbiology, and the steps that can be taken to make the predictions even more accurate, are discussed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge