Christina Röcke
Deep Generative Model-Based Generation of Synthetic Individual-Specific Brain MRI Segmentations
Apr 15, 2025Abstract:To the best of our knowledge, all existing methods that can generate synthetic brain magnetic resonance imaging (MRI) scans for a specific individual require detailed structural or volumetric information about the individual's brain. However, such brain information is often scarce, expensive, and difficult to obtain. In this paper, we propose the first approach capable of generating synthetic brain MRI segmentations -- specifically, 3D white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) segmentations -- for individuals using their easily obtainable and often readily available demographic, interview, and cognitive test information. Our approach features a novel deep generative model, CSegSynth, which outperforms existing prominent generative models, including conditional variational autoencoder (C-VAE), conditional generative adversarial network (C-GAN), and conditional latent diffusion model (C-LDM). We demonstrate the high quality of our synthetic segmentations through extensive evaluations. Also, in assessing the effectiveness of the individual-specific generation, we achieve superior volume prediction, with Pearson correlation coefficients reaching 0.80, 0.82, and 0.70 between the ground-truth WM, GM, and CSF volumes of test individuals and those volumes predicted based on generated individual-specific segmentations, respectively.
Eigenbehaviour as an Indicator of Cognitive Abilities
Oct 18, 2021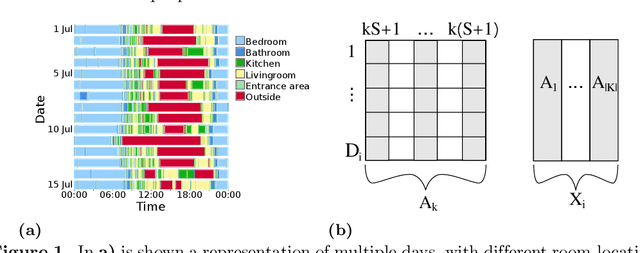

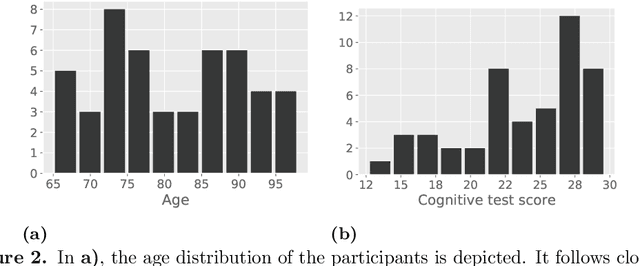
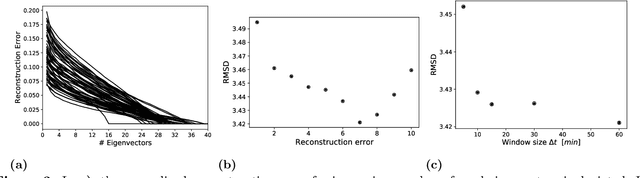
Abstract:With growing usage of machine learning algorithms and big data in health applications, digital biomarkers have become an important key feature to ensure the success of those applications. In this paper, we focus on one important use-case, the long-term continuous monitoring of the cognitive ability of older adults. The cognitive ability is a factor both for long-term monitoring of people living alone as well as an outcome in clinical studies. In this work, we propose a new digital biomarker for cognitive abilities based on location eigenbehaviour obtained from contactless ambient sensors. Indoor location information obtained from passive infrared sensors is used to build a location matrix covering several weeks of measurement. Based on the eigenvectors of this matrix, the reconstruction error is calculated for various numbers of used eigenvectors. The reconstruction error is used to predict cognitive ability scores collected at baseline, using linear regression. Additionally, classification of normal versus pathological cognition level is performed using a support-vector-machine. Prediction performance is strong for high levels of cognitive ability, but grows weaker for low levels of cognitive ability. Classification into normal versus pathological cognitive ability level reaches high accuracy with a AUC = 0.94. Due to the unobtrusive method of measurement based on contactless ambient sensors, this digital biomarker of cognitive ability is easily obtainable. The usage of the reconstruction error is a strong digital biomarker for the binary classification and, to a lesser extent, for more detailed prediction of interindividual differences in cognition.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge