Chengrui Li
A Hitchhiker's Guide to Poisson Gradient Estimation
Feb 03, 2026Abstract:Poisson-distributed latent variable models are widely used in computational neuroscience, but differentiating through discrete stochastic samples remains challenging. Two approaches address this: Exponential Arrival Time (EAT) simulation and Gumbel-SoftMax (GSM) relaxation. We provide the first systematic comparison of these methods, along with practical guidance for practitioners. Our main technical contribution is a modification to the EAT method that theoretically guarantees an unbiased first moment (exactly matching the firing rate), and reduces second-moment bias. We evaluate these methods on their distributional fidelity, gradient quality, and performance on two tasks: (1) variational autoencoders with Poisson latents, and (2) partially observable generalized linear models, where latent neural connectivity must be inferred from observed spike trains. Across all metrics, our modified EAT method exhibits better overall performance (often comparable to exact gradients), and substantially higher robustness to hyperparameter choices. Together, our results clarify the trade-offs between these methods and offer concrete recommendations for practitioners working with Poisson latent variable models.
A Disentangled Low-Rank RNN Framework for Uncovering Neural Connectivity and Dynamics
Nov 17, 2025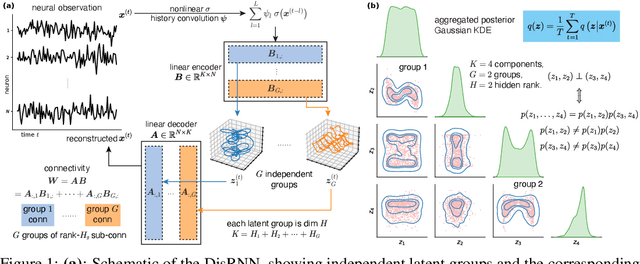
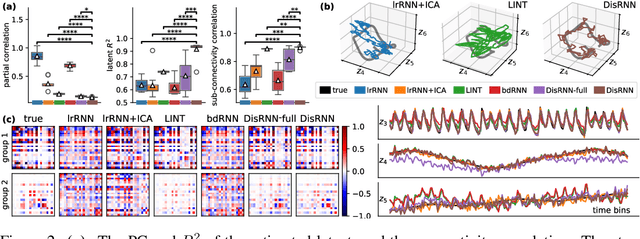
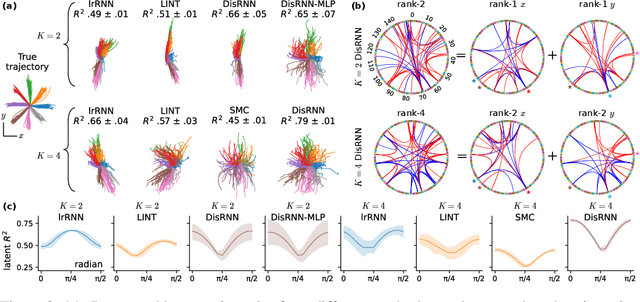
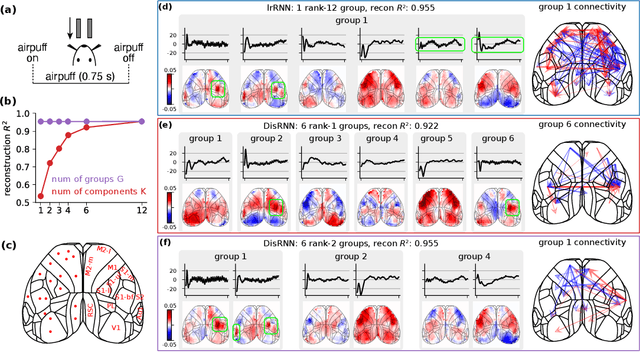
Abstract:Low-rank recurrent neural networks (lrRNNs) are a class of models that uncover low-dimensional latent dynamics underlying neural population activity. Although their functional connectivity is low-rank, it lacks disentanglement interpretations, making it difficult to assign distinct computational roles to different latent dimensions. To address this, we propose the Disentangled Recurrent Neural Network (DisRNN), a generative lrRNN framework that assumes group-wise independence among latent dynamics while allowing flexible within-group entanglement. These independent latent groups allow latent dynamics to evolve separately, but are internally rich for complex computation. We reformulate the lrRNN under a variational autoencoder (VAE) framework, enabling us to introduce a partial correlation penalty that encourages disentanglement between groups of latent dimensions. Experiments on synthetic, monkey M1, and mouse voltage imaging data show that DisRNN consistently improves the disentanglement and interpretability of learned neural latent trajectories in low-dimensional space and low-rank connectivity over baseline lrRNNs that do not encourage partial disentanglement.
Uncovering Semantic Selectivity of Latent Groups in Higher Visual Cortex with Mutual Information-Guided Diffusion
Oct 02, 2025Abstract:Understanding how neural populations in higher visual areas encode object-centered visual information remains a central challenge in computational neuroscience. Prior works have investigated representational alignment between artificial neural networks and the visual cortex. Nevertheless, these findings are indirect and offer limited insights to the structure of neural populations themselves. Similarly, decoding-based methods have quantified semantic features from neural populations but have not uncovered their underlying organizations. This leaves open a scientific question: "how feature-specific visual information is distributed across neural populations in higher visual areas, and whether it is organized into structured, semantically meaningful subspaces." To tackle this problem, we present MIG-Vis, a method that leverages the generative power of diffusion models to visualize and validate the visual-semantic attributes encoded in neural latent subspaces. Our method first uses a variational autoencoder to infer a group-wise disentangled neural latent subspace from neural populations. Subsequently, we propose a mutual information (MI)-guided diffusion synthesis procedure to visualize the specific visual-semantic features encoded by each latent group. We validate MIG-Vis on multi-session neural spiking datasets from the inferior temporal (IT) cortex of two macaques. The synthesized results demonstrate that our method identifies neural latent groups with clear semantic selectivity to diverse visual features, including object pose, inter-category transformations, and intra-class content. These findings provide direct, interpretable evidence of structured semantic representation in the higher visual cortex and advance our understanding of its encoding principles.
A Revisit of Total Correlation in Disentangled Variational Auto-Encoder with Partial Disentanglement
Feb 04, 2025
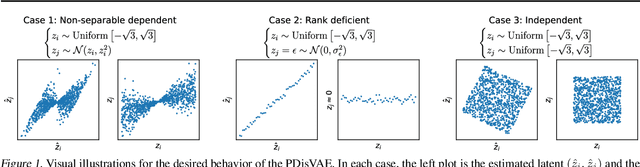
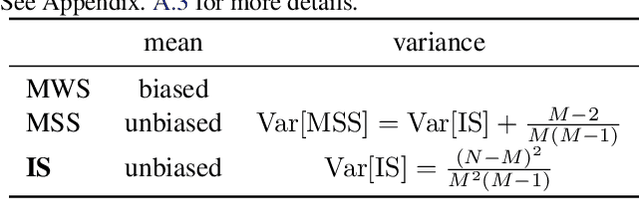
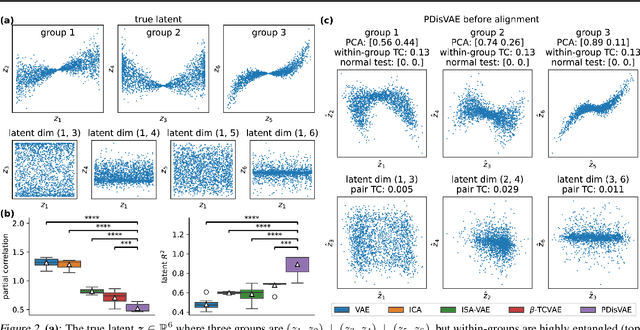
Abstract:A fully disentangled variational auto-encoder (VAE) aims to identify disentangled latent components from observations. However, enforcing full independence between all latent components may be too strict for certain datasets. In some cases, multiple factors may be entangled together in a non-separable manner, or a single independent semantic meaning could be represented by multiple latent components within a higher-dimensional manifold. To address such scenarios with greater flexibility, we develop the Partially Disentangled VAE (PDisVAE), which generalizes the total correlation (TC) term in fully disentangled VAEs to a partial correlation (PC) term. This framework can handle group-wise independence and can naturally reduce to either the standard VAE or the fully disentangled VAE. Validation through three synthetic experiments demonstrates the correctness and practicality of PDisVAE. When applied to real-world datasets, PDisVAE discovers valuable information that is difficult to find using fully disentangled VAEs, implying its versatility and effectiveness.
Exploring Behavior-Relevant and Disentangled Neural Dynamics with Generative Diffusion Models
Oct 12, 2024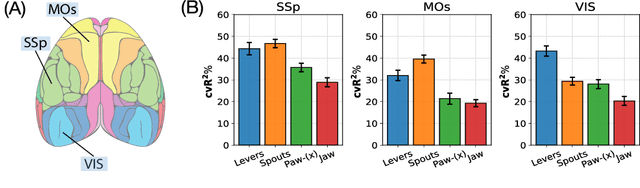
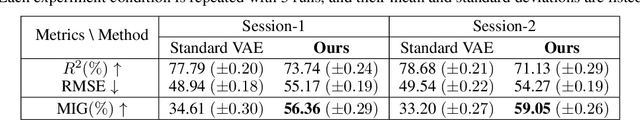
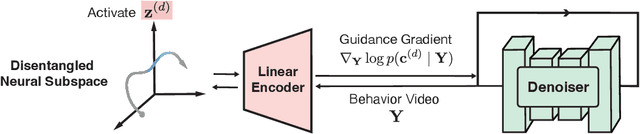
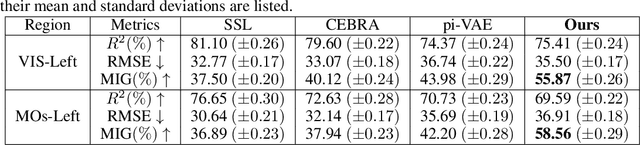
Abstract:Understanding the neural basis of behavior is a fundamental goal in neuroscience. Current research in large-scale neuro-behavioral data analysis often relies on decoding models, which quantify behavioral information in neural data but lack details on behavior encoding. This raises an intriguing scientific question: ``how can we enable in-depth exploration of neural representations in behavioral tasks, revealing interpretable neural dynamics associated with behaviors''. However, addressing this issue is challenging due to the varied behavioral encoding across different brain regions and mixed selectivity at the population level. To tackle this limitation, our approach, named ``BeNeDiff'', first identifies a fine-grained and disentangled neural subspace using a behavior-informed latent variable model. It then employs state-of-the-art generative diffusion models to synthesize behavior videos that interpret the neural dynamics of each latent factor. We validate the method on multi-session datasets containing widefield calcium imaging recordings across the dorsal cortex. Through guiding the diffusion model to activate individual latent factors, we verify that the neural dynamics of latent factors in the disentangled neural subspace provide interpretable quantifications of the behaviors of interest. At the same time, the neural subspace in BeNeDiff demonstrates high disentanglement and neural reconstruction quality.
Markovian Gaussian Process: A Universal State-Space Representation for Stationary Temporal Gaussian Process
Jun 29, 2024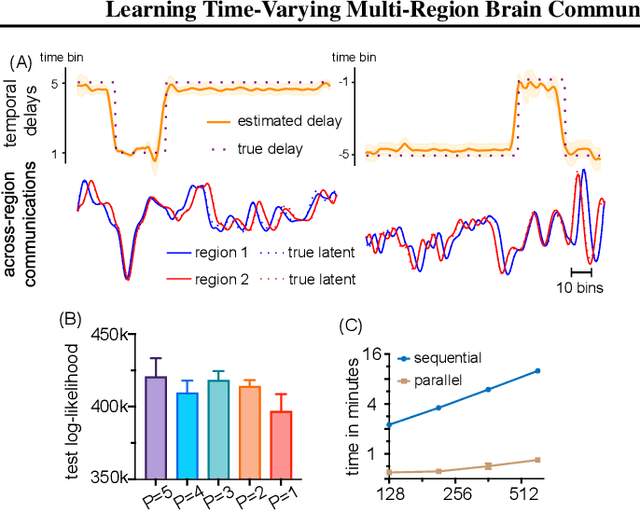

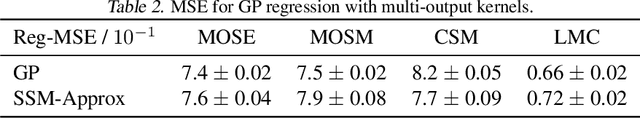
Abstract:Gaussian Processes (GPs) and Linear Dynamical Systems (LDSs) are essential time series and dynamic system modeling tools. GPs can handle complex, nonlinear dynamics but are computationally demanding, while LDSs offer efficient computation but lack the expressive power of GPs. To combine their benefits, we introduce a universal method that allows an LDS to mirror stationary temporal GPs. This state-space representation, known as the Markovian Gaussian Process (Markovian GP), leverages the flexibility of kernel functions while maintaining efficient linear computation. Unlike existing GP-LDS conversion methods, which require separability for most multi-output kernels, our approach works universally for single- and multi-output stationary temporal kernels. We evaluate our method by computing covariance, performing regression tasks, and applying it to a neuroscience application, demonstrating that our method provides an accurate state-space representation for stationary temporal GPs.
A Differentiable Partially Observable Generalized Linear Model with Forward-Backward Message Passing
Feb 07, 2024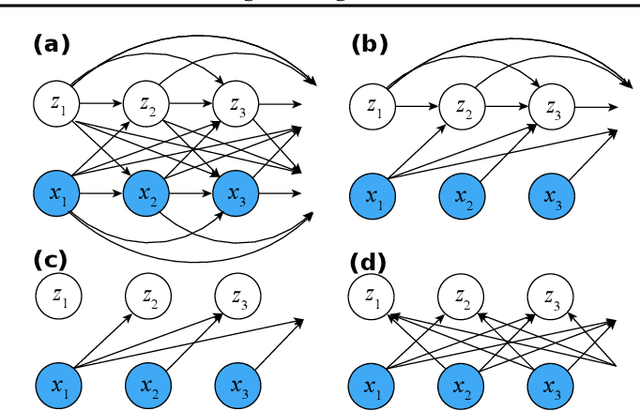
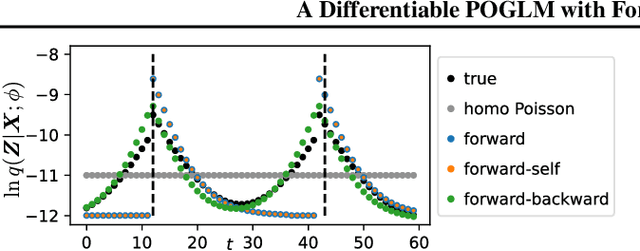
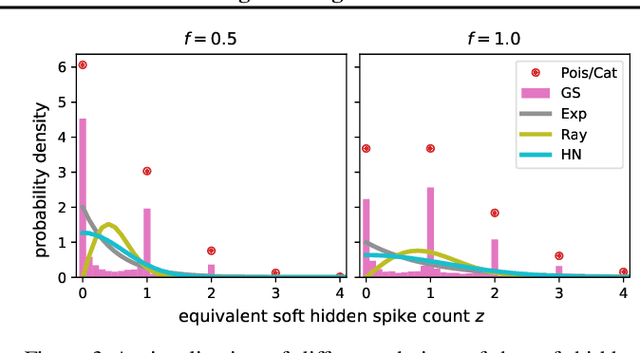
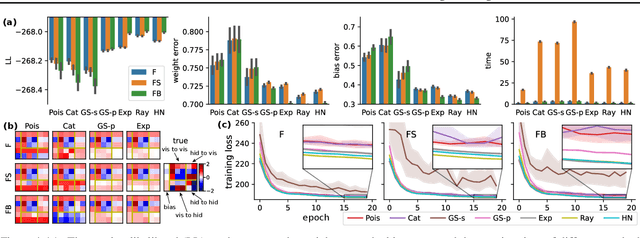
Abstract:The partially observable generalized linear model (POGLM) is a powerful tool for understanding neural connectivity under the assumption of existing hidden neurons. With spike trains only recorded from visible neurons, existing works use variational inference to learn POGLM meanwhile presenting the difficulty of learning this latent variable model. There are two main issues: (1) the sampled Poisson hidden spike count hinders the use of the pathwise gradient estimator in VI; and (2) the existing design of the variational model is neither expressive nor time-efficient, which further affects the performance. For (1), we propose a new differentiable POGLM, which enables the pathwise gradient estimator, better than the score function gradient estimator used in existing works. For (2), we propose the forward-backward message-passing sampling scheme for the variational model. Comprehensive experiments show that our differentiable POGLMs with our forward-backward message passing produce a better performance on one synthetic and two real-world datasets. Furthermore, our new method yields more interpretable parameters, underscoring its significance in neuroscience.
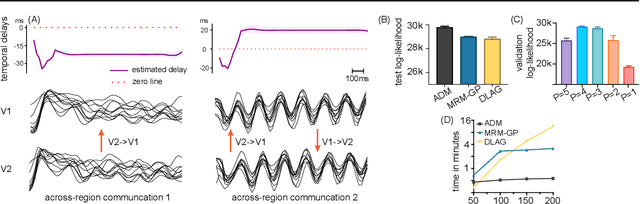
Multi-Region Markovian Gaussian Process: An Efficient Method to Discover Directional Communications Across Multiple Brain Regions
Feb 05, 2024Abstract:Studying the complex interactions between different brain regions is crucial in neuroscience. Various statistical methods have explored the latent communication across multiple brain regions. Two main categories are the Gaussian Process (GP) and Linear Dynamical System (LDS), each with unique strengths. The GP-based approach effectively discovers latent variables such as frequency bands and communication directions. Conversely, the LDS-based approach is computationally efficient but lacks powerful expressiveness in latent representation. In this study, we merge both methodologies by creating an LDS mirroring a multi-output GP, termed Multi-Region Markovian Gaussian Process (MRM-GP). Our work is the first to establish a connection between an LDS and a multi-output GP that explicitly models frequencies and phase delays within the latent space of neural recordings. Consequently, the model achieves a linear inference cost over time points and provides an interpretable low-dimensional representation, revealing communication directions across brain regions and separating oscillatory communications into different frequency bands.
Forward $χ^2$ Divergence Based Variational Importance Sampling
Nov 04, 2023



Abstract:Maximizing the log-likelihood is a crucial aspect of learning latent variable models, and variational inference (VI) stands as the commonly adopted method. However, VI can encounter challenges in achieving a high log-likelihood when dealing with complicated posterior distributions. In response to this limitation, we introduce a novel variational importance sampling (VIS) approach that directly estimates and maximizes the log-likelihood. VIS leverages the optimal proposal distribution, achieved by minimizing the forward $\chi^2$ divergence, to enhance log-likelihood estimation. We apply VIS to various popular latent variable models, including mixture models, variational auto-encoders, and partially observable generalized linear models. Results demonstrate that our approach consistently outperforms state-of-the-art baselines, both in terms of log-likelihood and model parameter estimation.
One-hot Generalized Linear Model for Switching Brain State Discovery
Oct 23, 2023



Abstract:Exposing meaningful and interpretable neural interactions is critical to understanding neural circuits. Inferred neural interactions from neural signals primarily reflect functional interactions. In a long experiment, subject animals may experience different stages defined by the experiment, stimuli, or behavioral states, and hence functional interactions can change over time. To model dynamically changing functional interactions, prior work employs state-switching generalized linear models with hidden Markov models (i.e., HMM-GLMs). However, we argue they lack biological plausibility, as functional interactions are shaped and confined by the underlying anatomical connectome. Here, we propose a novel prior-informed state-switching GLM. We introduce both a Gaussian prior and a one-hot prior over the GLM in each state. The priors are learnable. We will show that the learned prior should capture the state-constant interaction, shedding light on the underlying anatomical connectome and revealing more likely physical neuron interactions. The state-dependent interaction modeled by each GLM offers traceability to capture functional variations across multiple brain states. Our methods effectively recover true interaction structures in simulated data, achieve the highest predictive likelihood with real neural datasets, and render interaction structures and hidden states more interpretable when applied to real neural data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge