Chau Hung Lee
Segmenting Medical Images with Limited Data
Jul 12, 2024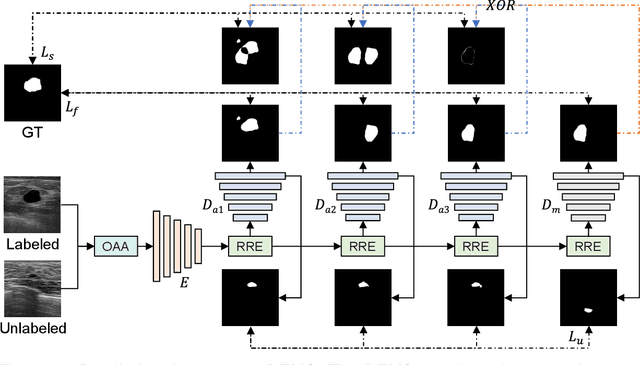
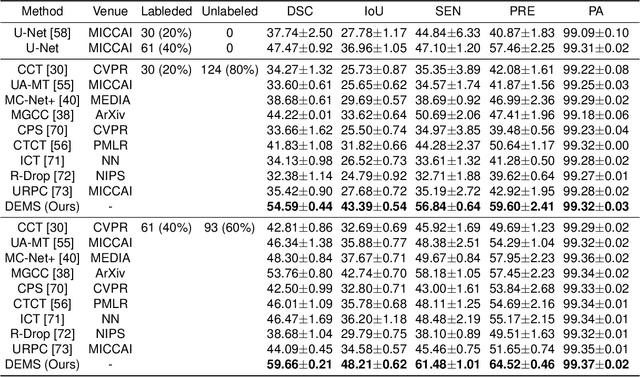
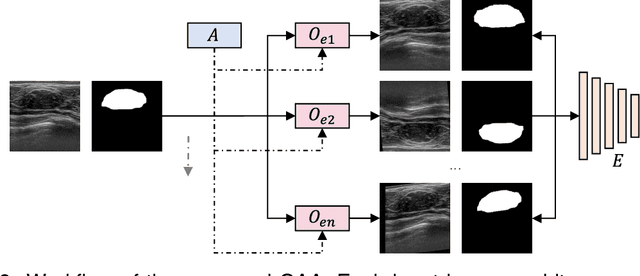
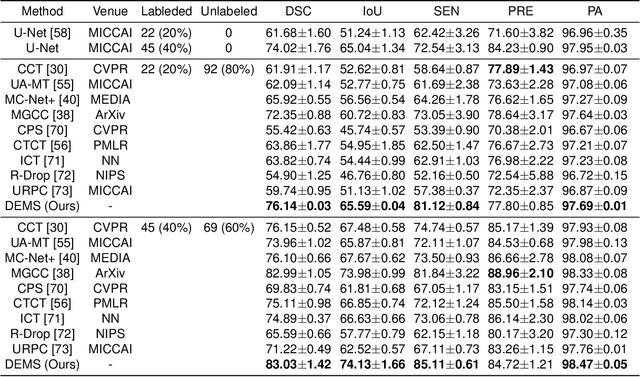
Abstract:While computer vision has proven valuable for medical image segmentation, its application faces challenges such as limited dataset sizes and the complexity of effectively leveraging unlabeled images. To address these challenges, we present a novel semi-supervised, consistency-based approach termed the data-efficient medical segmenter (DEMS). The DEMS features an encoder-decoder architecture and incorporates the developed online automatic augmenter (OAA) and residual robustness enhancement (RRE) blocks. The OAA augments input data with various image transformations, thereby diversifying the dataset to improve the generalization ability. The RRE enriches feature diversity and introduces perturbations to create varied inputs for different decoders, thereby providing enhanced variability. Moreover, we introduce a sensitive loss to further enhance consistency across different decoders and stabilize the training process. Extensive experimental results on both our own and three public datasets affirm the effectiveness of DEMS. Under extreme data shortage scenarios, our DEMS achieves 16.85\% and 10.37\% improvement in dice score compared with the U-Net and top-performed state-of-the-art method, respectively. Given its superior data efficiency, DEMS could present significant advancements in medical segmentation under small data regimes. The project homepage can be accessed at https://github.com/NUS-Tim/DEMS.
Semi-supervised classification of medical ultrasound images based on generative adversarial network
Mar 11, 2022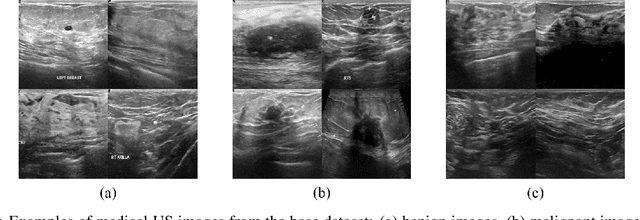
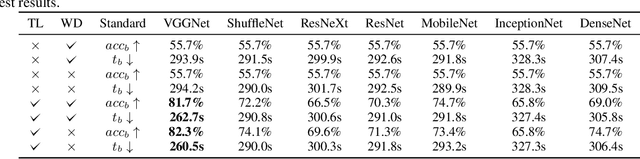
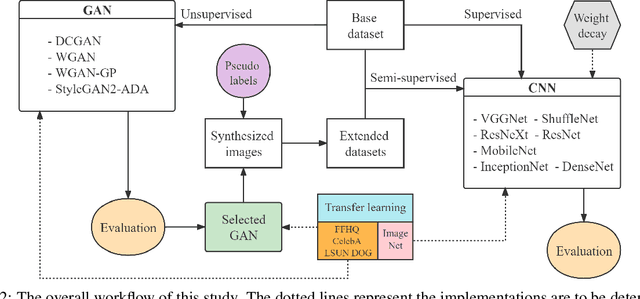
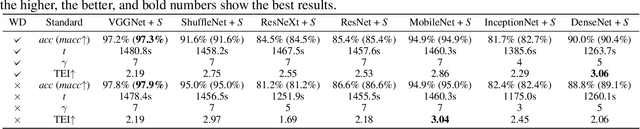
Abstract:Medical ultrasound (US) is one of the most widely used imaging modalities in clinical practice. However, its use presents unique challenges such as variable imaging quality. Deep learning (DL) can be used as an advanced medical US images analysis tool, while the performance of the DL model is greatly limited by the scarcity of big datasets. Here, we develop semi-supervised classification enhancement (SSCE) structures by constructing seven convolutional neural network (CNN) models and one of the most state-of-the-art generative adversarial network (GAN) models, StyleGAN2-ADA, to address this problem. A breast cancer dataset with 780 images is used as our base dataset. The results show that our SSCE structures obtain an accuracy of up to 97.9%, showing a maximum 21.6% improvement compared with utilizing CNN models alone and outperforming the previous methods using the same dataset by up to 23.9%. We believe our proposed state-of-the-art method can be regarded as a potential auxiliary tool for on-the-fly diagnoses of medical US images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge