Carri Glide-Hurst
Deterministic Medical Image Translation via High-fidelity Brownian Bridges
Mar 28, 2025Abstract:Recent studies have shown that diffusion models produce superior synthetic images when compared to Generative Adversarial Networks (GANs). However, their outputs are often non-deterministic and lack high fidelity to the ground truth due to the inherent randomness. In this paper, we propose a novel High-fidelity Brownian bridge model (HiFi-BBrg) for deterministic medical image translations. Our model comprises two distinct yet mutually beneficial mappings: a generation mapping and a reconstruction mapping. The Brownian bridge training process is guided by the fidelity loss and adversarial training in the reconstruction mapping. This ensures that translated images can be accurately reversed to their original forms, thereby achieving consistent translations with high fidelity to the ground truth. Our extensive experiments on multiple datasets show HiFi-BBrg outperforms state-of-the-art methods in multi-modal image translation and multi-image super-resolution.
Conditional diffusion model with spatial attention and latent embedding for medical image segmentation
Feb 10, 2025Abstract:Diffusion models have been used extensively for high quality image and video generation tasks. In this paper, we propose a novel conditional diffusion model with spatial attention and latent embedding (cDAL) for medical image segmentation. In cDAL, a convolutional neural network (CNN) based discriminator is used at every time-step of the diffusion process to distinguish between the generated labels and the real ones. A spatial attention map is computed based on the features learned by the discriminator to help cDAL generate more accurate segmentation of discriminative regions in an input image. Additionally, we incorporated a random latent embedding into each layer of our model to significantly reduce the number of training and sampling time-steps, thereby making it much faster than other diffusion models for image segmentation. We applied cDAL on 3 publicly available medical image segmentation datasets (MoNuSeg, Chest X-ray and Hippocampus) and observed significant qualitative and quantitative improvements with higher Dice scores and mIoU over the state-of-the-art algorithms. The source code is publicly available at https://github.com/Hejrati/cDAL/.
Modality-Agnostic Learning for Medical Image Segmentation Using Multi-modality Self-distillation
Jun 06, 2023Abstract:Medical image segmentation of tumors and organs at risk is a time-consuming yet critical process in the clinic that utilizes multi-modality imaging (e.g, different acquisitions, data types, and sequences) to increase segmentation precision. In this paper, we propose a novel framework, Modality-Agnostic learning through Multi-modality Self-dist-illation (MAG-MS), to investigate the impact of input modalities on medical image segmentation. MAG-MS distills knowledge from the fusion of multiple modalities and applies it to enhance representation learning for individual modalities. Thus, it provides a versatile and efficient approach to handle limited modalities during testing. Our extensive experiments on benchmark datasets demonstrate the high efficiency of MAG-MS and its superior segmentation performance than current state-of-the-art methods. Furthermore, using MAG-MS, we provide valuable insight and guidance on selecting input modalities for medical image segmentation tasks.
Dual self-distillation of U-shaped networks for 3D medical image segmentation
Jun 05, 2023



Abstract:U-shaped networks and its variants have demonstrated exceptional results for medical image segmentation. In this paper, we propose a novel dual self-distillation (DSD) framework for U-shaped networks for 3D medical image segmentation. DSD distills knowledge from the ground-truth segmentation labels to the decoder layers and also between the encoder and decoder layers of a single U-shaped network. DSD is a generalized training strategy that could be attached to the backbone architecture of any U-shaped network to further improve its segmentation performance. We attached DSD on two state-of-the-art U-shaped backbones, and extensive experiments on two public 3D medical image segmentation datasets (cardiac substructure and brain tumor) demonstrated significant improvement over those backbones. On average, after attaching DSD to the U-shaped backbones, we observed an improvement of 4.25% and 3.15% in Dice similarity score for cardiac substructure and brain tumor segmentation respectively.
SA-GAN: Structure-Aware Generative Adversarial Network for Shape-Preserving Synthetic CT Generation
May 14, 2021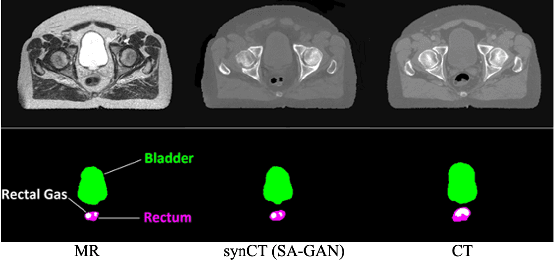
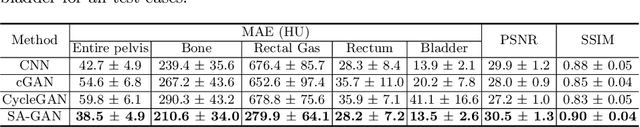
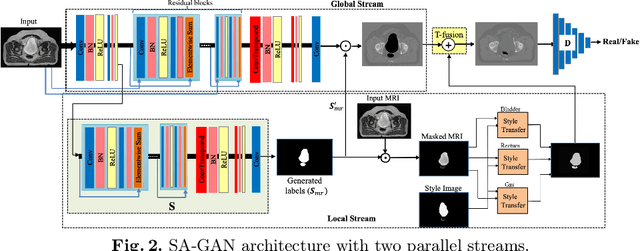
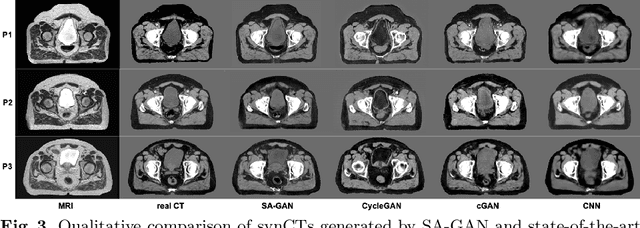
Abstract:In medical image synthesis, model training could be challenging due to the inconsistencies between images of different modalities even with the same patient, typically caused by internal status/tissue changes as different modalities are usually obtained at a different time. This paper proposes a novel deep learning method, Structure-aware Generative Adversarial Network (SA-GAN), that preserves the shapes and locations of in-consistent structures when generating medical images. SA-GAN is employed to generate synthetic computed tomography (synCT) images from magnetic resonance imaging (MRI) with two parallel streams: the global stream translates the input from the MRI to the CT domain while the local stream automatically segments the inconsistent organs, maintains their locations and shapes in MRI, and translates the organ intensities to CT. Through extensive experiments on a pelvic dataset, we demonstrate that SA-GAN provides clinically acceptable accuracy on both synCTs and organ segmentation and supports MR-only treatment planning in disease sites with internal organ status changes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge