Can Fahrettin Koyuncu
Leveraging Weak Supervision for Cell Localization in Digital Pathology Using Multitask Learning and Consistency Loss
Dec 19, 2024Abstract:Cell detection and segmentation are integral parts of automated systems in digital pathology. Encoder-decoder networks have emerged as a promising solution for these tasks. However, training of these networks has typically required full boundary annotations of cells, which are labor-intensive and difficult to obtain on a large scale. However, in many applications, such as cell counting, weaker forms of annotations--such as point annotations or approximate cell counts--can provide sufficient supervision for training. This study proposes a new mixed-supervision approach for training multitask networks in digital pathology by incorporating cell counts derived from the eyeballing process--a quick visual estimation method commonly used by pathologists. This study has two main contributions: (1) It proposes a mixed-supervision strategy for digital pathology that utilizes cell counts obtained by eyeballing as an auxiliary supervisory signal to train a multitask network for the first time. (2) This multitask network is designed to concurrently learn the tasks of cell counting and cell localization, and this study introduces a consistency loss that regularizes training by penalizing inconsistencies between the predictions of these two tasks. Our experiments on two datasets of hematoxylin-eosin stained tissue images demonstrate that the proposed approach effectively utilizes the weakest form of annotation, improving performance when stronger annotations are limited. These results highlight the potential of integrating eyeballing-derived ground truths into the network training, reducing the need for resource-intensive annotations.
DeepDistance: A Multi-task Deep Regression Model for Cell Detection in Inverted Microscopy Images
Aug 29, 2019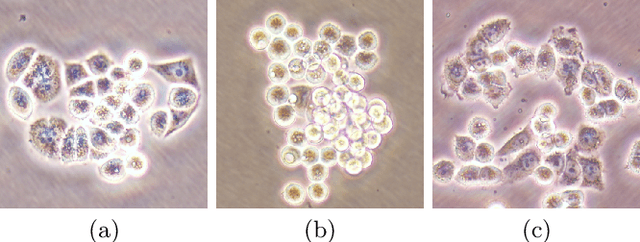
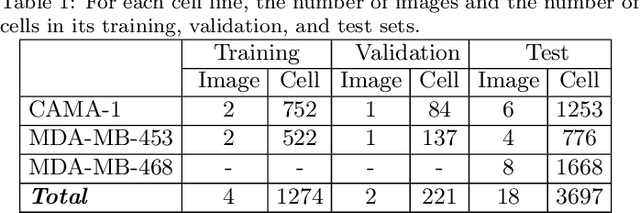
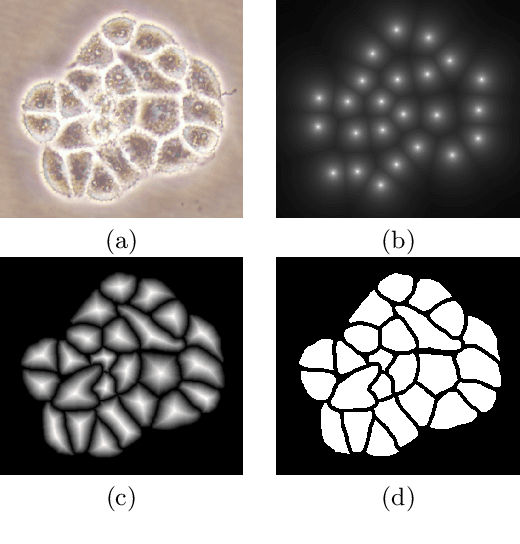
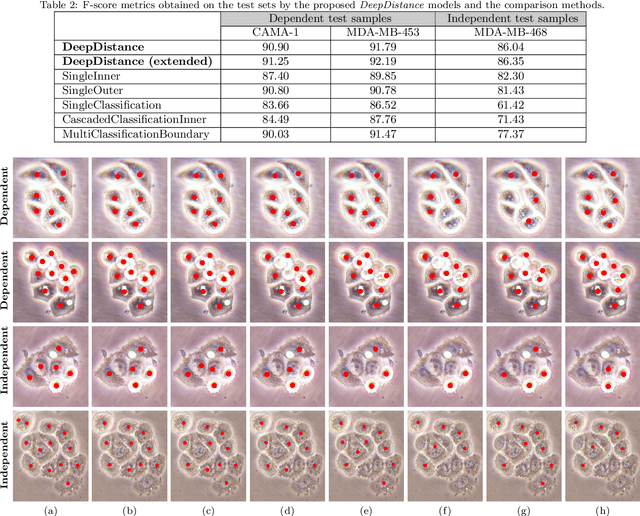
Abstract:This paper presents a new deep regression model, which we call DeepDistance, for cell detection in images acquired with inverted microscopy. This model considers cell detection as a task of finding most probable locations that suggest cell centers in an image. It represents this main task with a regression task of learning an inner distance metric. However, different than the previously reported regression based methods, the DeepDistance model proposes to approach its learning as a multi-task regression problem where multiple tasks are learned by using shared feature representations. To this end, it defines a secondary metric, normalized outer distance, to represent a different aspect of the problem and proposes to define its learning as complementary to the main cell detection task. In order to learn these two complementary tasks more effectively, the DeepDistance model designs a fully convolutional network (FCN) with a shared encoder path and end-to-end trains this FCN to concurrently learn the tasks in parallel. DeepDistance uses the inner distances estimated by this FCN in a detection algorithm to locate individual cells in a given image. For further performance improvement on the main task, this paper also presents an extended version of the DeepDistance model. This extended model includes an auxiliary classification task and learns it in parallel to the two regression tasks by sharing feature representations with them. Our experiments on three different human cell lines reveal that the proposed multi-task learning models, the DeepDistance model and its extended version, successfully identify cell locations, even for the cell line that was not used in training, and improve the results of the previous deep learning methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge