Cagla Deniz Bahadir
Characterizing the Features of Mitotic Figures Using a Conditional Diffusion Probabilistic Model
Oct 05, 2023



Abstract:Mitotic figure detection in histology images is a hard-to-define, yet clinically significant task, where labels are generated with pathologist interpretations and where there is no ``gold-standard'' independent ground-truth. However, it is well-established that these interpretation based labels are often unreliable, in part, due to differences in expertise levels and human subjectivity. In this paper, our goal is to shed light on the inherent uncertainty of mitosis labels and characterize the mitotic figure classification task in a human interpretable manner. We train a probabilistic diffusion model to synthesize patches of cell nuclei for a given mitosis label condition. Using this model, we can then generate a sequence of synthetic images that correspond to the same nucleus transitioning into the mitotic state. This allows us to identify different image features associated with mitosis, such as cytoplasm granularity, nuclear density, nuclear irregularity and high contrast between the nucleus and the cell body. Our approach offers a new tool for pathologists to interpret and communicate the features driving the decision to recognize a mitotic figure.
Learning-based Optimization of the Under-sampling Pattern in MRI
Jan 07, 2019
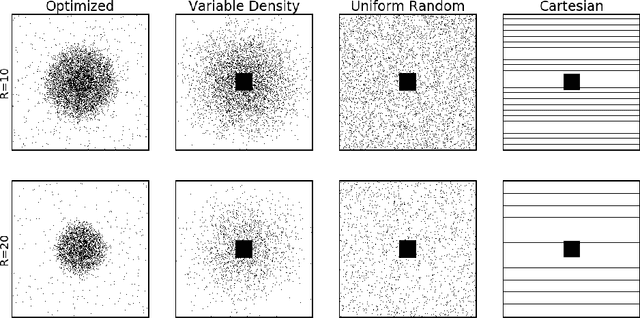
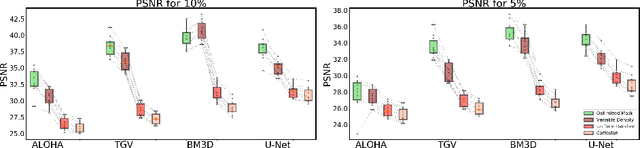

Abstract:Acquisition of Magnetic Resonance Imaging (MRI) scans can be accelerated by under-sampling in k-space (i.e., the Fourier domain). In this paper, we consider the problem of optimizing the sub-sampling pattern in a data-driven fashion. Since the reconstruction model's performance depends on the sub-sampling pattern, we combine the two problems. For a given sparsity constraint, our method optimizes the sub-sampling pattern and reconstruction model, using an end-to-end learning strategy. Our algorithm learns from full-resolution data that are under-sampled retrospectively, yielding a sub-sampling pattern and reconstruction model that are customized to the type of images represented in the training data. The proposed method, which we call LOUPE (Learning-based Optimization of the Under-sampling PattErn), was implemented by modifying a U-Net, a widely-used convolutional neural network architecture, that we append with the forward model that encodes the under-sampling process. Our experiments with T1-weighted structural brain MRI scans show that the optimized sub-sampling pattern can yield significantly more accurate reconstructions compared to standard random uniform, variable density or equispaced under-sampling schemes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge