Caetano Traina Jr.
A superpixel-driven deep learning approach for the analysis of dermatological wounds
Sep 20, 2019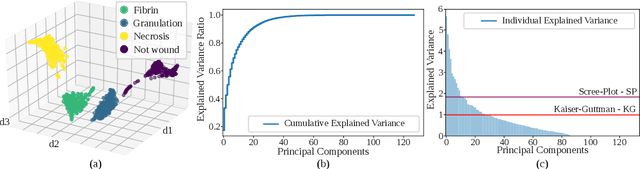
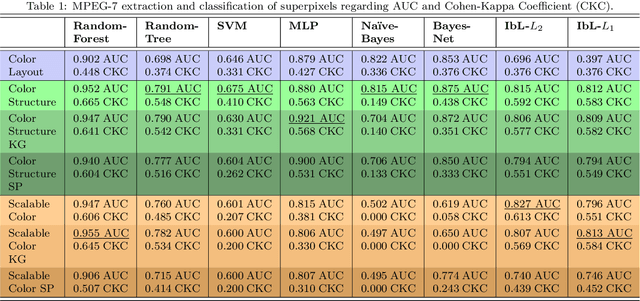
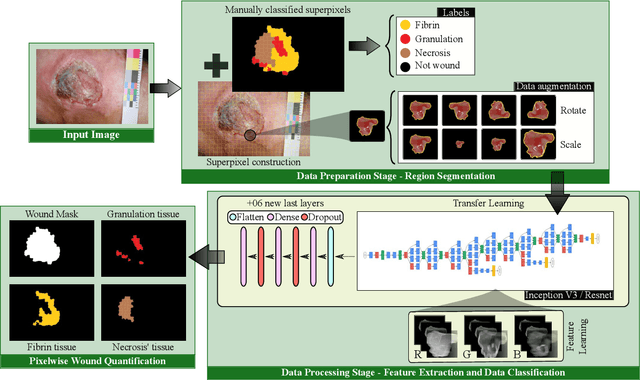
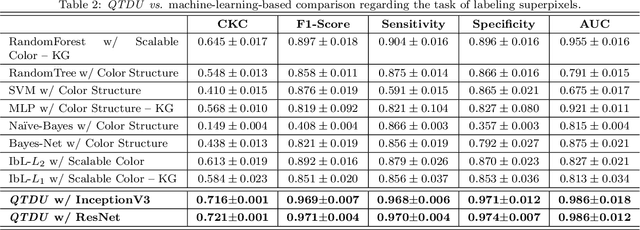
Abstract:Background. The image-based identification of distinct tissues within dermatological wounds enhances patients' care since it requires no intrusive evaluations. This manuscript presents an approach, we named QTDU, that combines deep learning models with superpixel-driven segmentation methods for assessing the quality of tissues from dermatological ulcers. Method. QTDU consists of a three-stage pipeline for the obtaining of ulcer segmentation, tissues' labeling, and wounded area quantification. We set up our approach by using a real and annotated set of dermatological ulcers for training several deep learning models to the identification of ulcered superpixels. Results. Empirical evaluations on 179,572 superpixels divided into four classes showed QTDU accurately spot wounded tissues (AUC = 0.986, sensitivity = 0.97, and specificity = 0.974) and outperformed machine-learning approaches in up to 8.2% regarding F1-Score through fine-tuning of a ResNet-based model. Last, but not least, experimental evaluations also showed QTDU correctly quantified wounded tissue areas within a 0.089 Mean Absolute Error ratio. Conclusions. Results indicate QTDU effectiveness for both tissue segmentation and wounded area quantification tasks. When compared to existing machine-learning approaches, the combination of superpixels and deep learning models outperformed the competitors within strong significant levels.
3DBGrowth: volumetric vertebrae segmentation and reconstruction in magnetic resonance imaging
Jul 08, 2019



Abstract:Segmentation of medical images is critical for making several processes of analysis and classification more reliable. With the growing number of people presenting back pain and related problems, the semi-automatic segmentation and 3D reconstruction of vertebral bodies became even more important to support decision making. A 3D reconstruction allows a fast and objective analysis of each vertebrae condition, which may play a major role in surgical planning and evaluation of suitable treatments. In this paper, we propose 3DBGrowth, which develops a 3D reconstruction over the efficient Balanced Growth method for 2D images. We also take advantage of the slope coefficient from the annotation time to reduce the total number of annotated slices, reducing the time spent on manual annotation. We show experimental results on a representative dataset with 17 MRI exams demonstrating that our approach significantly outperforms the competitors and, on average, only 37% of the total slices with vertebral body content must be annotated without losing performance/accuracy. Compared to the state-of-the-art methods, we have achieved a Dice Score gain of over 5% with comparable processing time. Moreover, 3DBGrowth works well with imprecise seed points, which reduces the time spent on manual annotation by the specialist.
* This is a pre-print of an article published in Computer-Based Medical Systems. The final authenticated version is available online at: https://doi.org/10.1109/CBMS.2019.00091
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge