Bruno Godefroy
Modeling Drug-Disease Relations with Linguistic and Knowledge Graph Constraints
Mar 31, 2019



Abstract:FDA drug labels are rich sources of information about drugs and drug-disease relations, but their complexity makes them challenging texts to analyze in isolation. To overcome this, we situate these labels in two health knowledge graphs: one built from precise structured information about drugs and diseases, and another built entirely from a database of clinical narrative texts using simple heuristic methods. We show that Probabilistic Soft Logic models defined over these graphs are superior to text-only and relation-only variants, and that the clinical narratives graph delivers exceptional results with little manual effort. Finally, we release a new dataset of drug labels with annotations for five distinct drug-disease relations.
Effective Feature Representation for Clinical Text Concept Extraction
Oct 31, 2018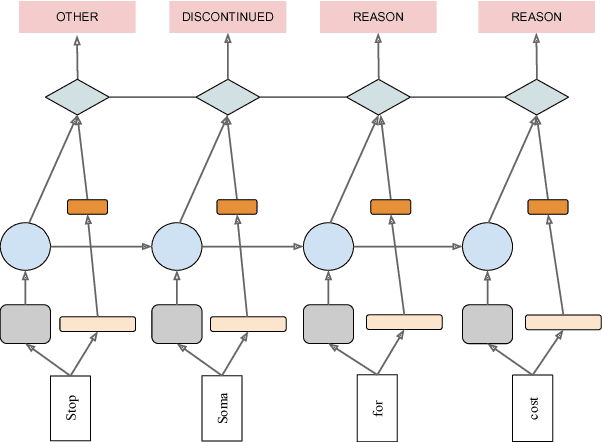
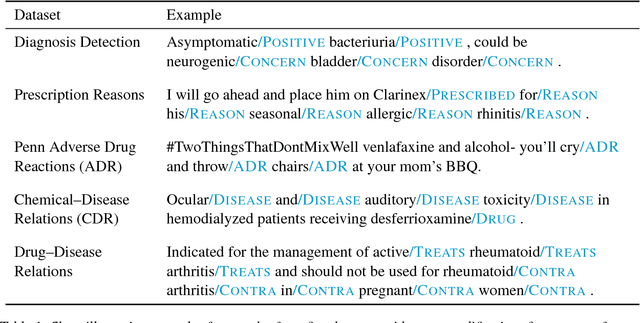
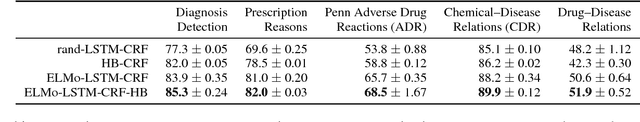
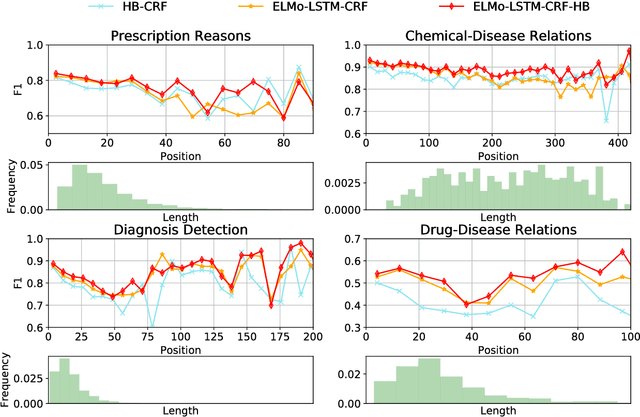
Abstract:Crucial information about the practice of healthcare is recorded only in free-form text, which creates an enormous opportunity for high-impact NLP. However, annotated healthcare datasets tend to be small and expensive to obtain, which raises the question of how to make maximally efficient uses of the available data. To this end, we develop an LSTM-CRF model for combining unsupervised word representations and hand-built feature representations derived from publicly available healthcare ontologies. We show that this combined model yields superior performance on five datasets of diverse kinds of healthcare text (clinical, social, scientific, commercial). Each involves the labeling of complex, multi-word spans that pick out different healthcare concepts. We also introduce a new labeled dataset for identifying the treatment relations between drugs and diseases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge