Brian Pollack
Explaining the Black-box Smoothly- A Counterfactual Approach
Jan 11, 2021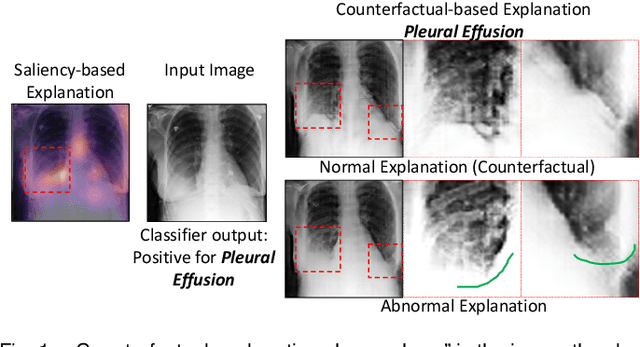
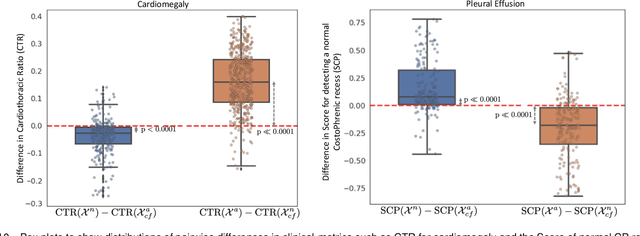
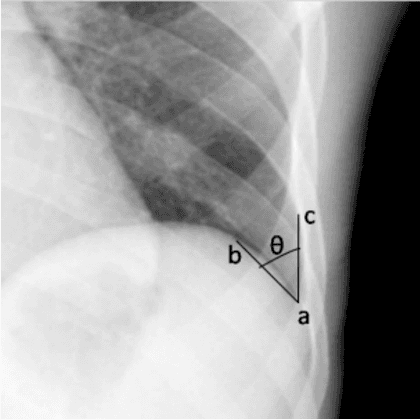
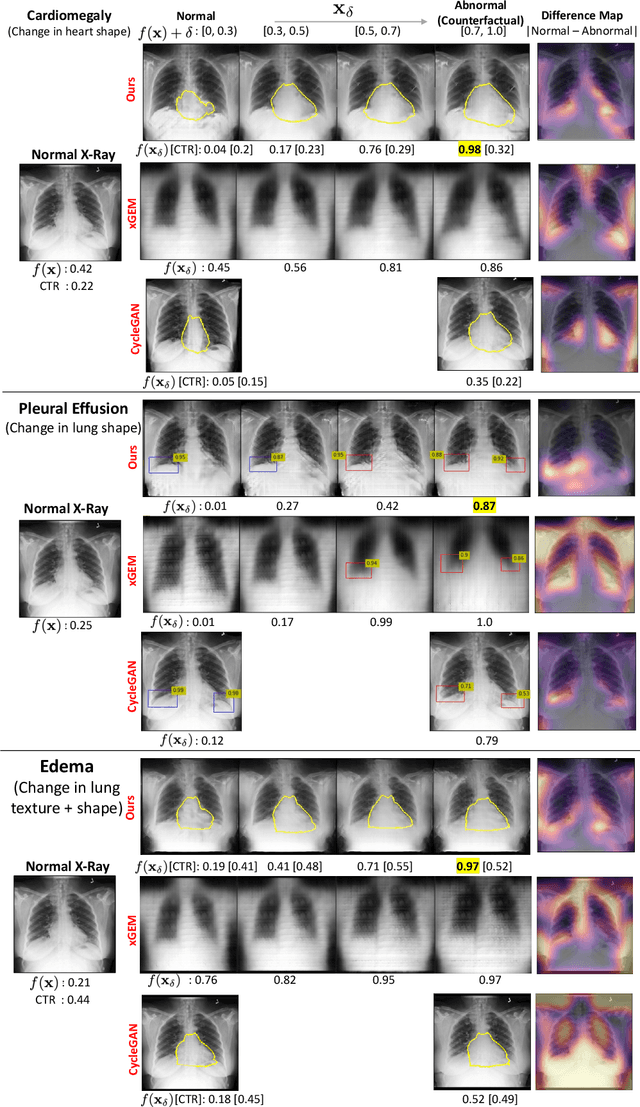
Abstract:We propose a BlackBox \emph{Counterfactual Explainer} that is explicitly developed for medical imaging applications. Classical approaches (e.g. saliency maps) assessing feature importance do not explain \emph{how} and \emph{why} variations in a particular anatomical region is relevant to the outcome, which is crucial for transparent decision making in healthcare application. Our framework explains the outcome by gradually \emph{exaggerating} the semantic effect of the given outcome label. Given a query input to a classifier, Generative Adversarial Networks produce a progressive set of perturbations to the query image that gradually changes the posterior probability from its original class to its negation. We design the loss function to ensure that essential and potentially relevant details, such as support devices, are preserved in the counterfactually generated images. We provide an extensive evaluation of different classification tasks on the chest X-Ray images. Our experiments show that a counterfactually generated visual explanation is consistent with the disease's clinical relevant measurements, both quantitatively and qualitatively.
Explanation by Progressive Exaggeration
Nov 05, 2019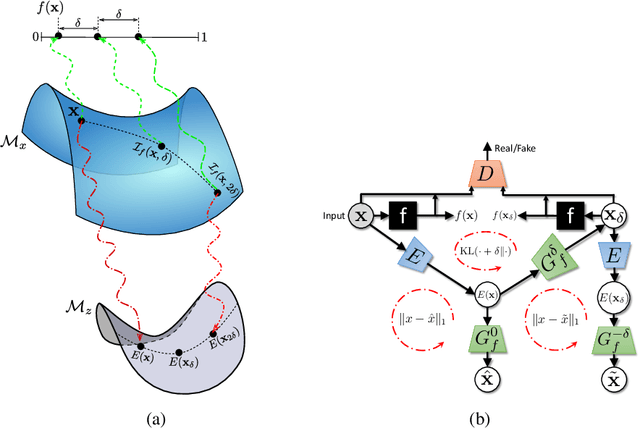
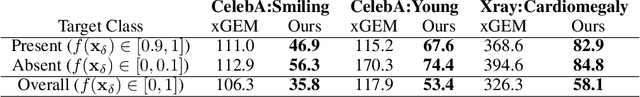
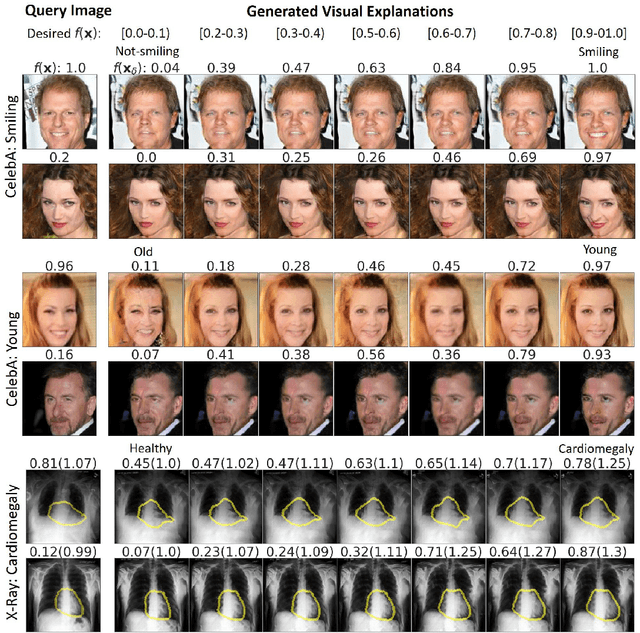

Abstract:As machine learning methods see greater adoption and implementation in high stakes applications such as medical image diagnosis, the need for model interpretability and explanation has become more critical. Classical approaches that assess feature importance (e.g. saliency maps) do not explain how and why a particular region of an image is relevant to the prediction. We propose a method that explains the outcome of a classification black-box by gradually exaggerating the semantic effect of a given class. Given a query input to a classifier, our method produces a progressive set of plausible variations of that query, which gradually changes the posterior probability from its original class to its negation. These counter-factually generated samples preserve features unrelated to the classification decision, such that a user can employ our method as a "tuning knob" to traverse a data manifold while crossing the decision boundary. Our method is model agnostic and only requires the output value and gradient of the predictor with respect to its input.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge