Bojana Bašaragin
Improving customer service with automatic topic detection in user emails
Feb 26, 2025Abstract:This study introduces a novel Natural Language Processing pipeline that enhances customer service efficiency at Telekom Srbija, a leading Serbian telecommunications company, through automated email topic detection and labelling. Central to the pipeline is BERTopic, a modular architecture that allows unsupervised topic modelling. After a series of preprocessing and post-processing steps, we assign one of 12 topics and several additional labels to incoming emails, allowing customer service to filter and access them through a custom-made application. The model's performance was evaluated by assessing the speed and correctness of the automatically assigned topics across a test dataset of 100 customer emails. The pipeline shows broad applicability across languages, particularly for those that are low-resourced and morphologically rich. The system now operates in the company's production environment, streamlining customer service operations through automated email classification.
Scientific QA System with Verifiable Answers
Jul 16, 2024


Abstract:In this paper, we introduce the VerifAI project, a pioneering open-source scientific question-answering system, designed to provide answers that are not only referenced but also automatically vetted and verifiable. The components of the system are (1) an Information Retrieval system combining semantic and lexical search techniques over scientific papers (PubMed), (2) a Retrieval-Augmented Generation (RAG) module using fine-tuned generative model (Mistral 7B) and retrieved articles to generate claims with references to the articles from which it was derived, and (3) a Verification engine, based on a fine-tuned DeBERTa and XLM-RoBERTa models on Natural Language Inference task using SciFACT dataset. The verification engine cross-checks the generated claim and the article from which the claim was derived, verifying whether there may have been any hallucinations in generating the claim. By leveraging the Information Retrieval and RAG modules, Verif.ai excels in generating factual information from a vast array of scientific sources. At the same time, the Verification engine rigorously double-checks this output, ensuring its accuracy and reliability. This dual-stage process plays a crucial role in acquiring and confirming factual information, significantly enhancing the information landscape. Our methodology could significantly enhance scientists' productivity, concurrently fostering trust in applying generative language models within scientific domains, where hallucinations and misinformation are unacceptable.
How do you know that? Teaching Generative Language Models to Reference Answers to Biomedical Questions
Jul 06, 2024



Abstract:Large language models (LLMs) have recently become the leading source of answers for users' questions online. Despite their ability to offer eloquent answers, their accuracy and reliability can pose a significant challenge. This is especially true for sensitive domains such as biomedicine, where there is a higher need for factually correct answers. This paper introduces a biomedical retrieval-augmented generation (RAG) system designed to enhance the reliability of generated responses. The system is based on a fine-tuned LLM for the referenced question-answering, where retrieved relevant abstracts from PubMed are passed to LLM's context as input through a prompt. Its output is an answer based on PubMed abstracts, where each statement is referenced accordingly, allowing the users to verify the answer. Our retrieval system achieves an absolute improvement of 23% compared to the PubMed search engine. Based on the manual evaluation on a small sample, our fine-tuned LLM component achieves comparable results to GPT-4 Turbo in referencing relevant abstracts. We make the dataset used to fine-tune the models and the fine-tuned models based on Mistral-7B-instruct-v0.1 and v0.2 publicly available.
Multilingual transformer and BERTopic for short text topic modeling: The case of Serbian
Feb 05, 2024Abstract:This paper presents the results of the first application of BERTopic, a state-of-the-art topic modeling technique, to short text written in a morphologi-cally rich language. We applied BERTopic with three multilingual embed-ding models on two levels of text preprocessing (partial and full) to evalu-ate its performance on partially preprocessed short text in Serbian. We also compared it to LDA and NMF on fully preprocessed text. The experiments were conducted on a dataset of tweets expressing hesitancy toward COVID-19 vaccination. Our results show that with adequate parameter setting, BERTopic can yield informative topics even when applied to partially pre-processed short text. When the same parameters are applied in both prepro-cessing scenarios, the performance drop on partially preprocessed text is minimal. Compared to LDA and NMF, judging by the keywords, BERTopic offers more informative topics and gives novel insights when the number of topics is not limited. The findings of this paper can be significant for re-searchers working with other morphologically rich low-resource languages and short text.
De-identification of clinical free text using natural language processing: A systematic review of current approaches
Nov 28, 2023Abstract:Background: Electronic health records (EHRs) are a valuable resource for data-driven medical research. However, the presence of protected health information (PHI) makes EHRs unsuitable to be shared for research purposes. De-identification, i.e. the process of removing PHI is a critical step in making EHR data accessible. Natural language processing has repeatedly demonstrated its feasibility in automating the de-identification process. Objectives: Our study aims to provide systematic evidence on how the de-identification of clinical free text has evolved in the last thirteen years, and to report on the performances and limitations of the current state-of-the-art systems. In addition, we aim to identify challenges and potential research opportunities in this field. Methods: A systematic search in PubMed, Web of Science and the DBLP was conducted for studies published between January 2010 and February 2023. Titles and abstracts were examined to identify the relevant studies. Selected studies were then analysed in-depth, and information was collected on de-identification methodologies, data sources, and measured performance. Results: A total of 2125 publications were identified for the title and abstract screening. 69 studies were found to be relevant. Machine learning (37 studies) and hybrid (26 studies) approaches are predominant, while six studies relied only on rules. Majority of the approaches were trained and evaluated on public corpora. The 2014 i2b2/UTHealth corpus is the most frequently used (36 studies), followed by the 2006 i2b2 (18 studies) and 2016 CEGS N-GRID (10 studies) corpora.
A transformer-based method for zero and few-shot biomedical named entity recognition
May 12, 2023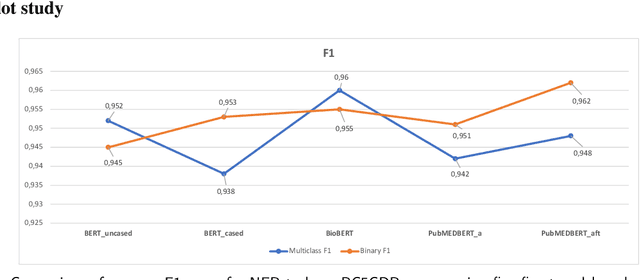
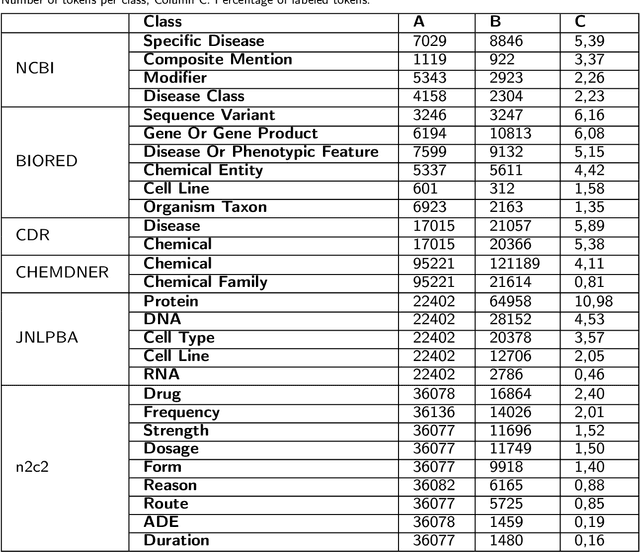

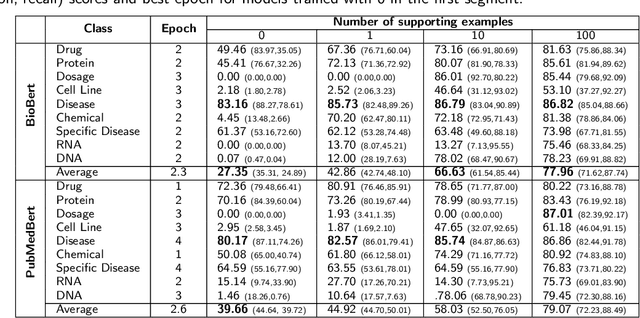
Abstract:Supervised named entity recognition (NER) in the biomedical domain is dependent on large sets of annotated texts with the given named entities, whose creation can be time-consuming and expensive. Furthermore, the extraction of new entities often requires conducting additional annotation tasks and retraining the model. To address these challenges, this paper proposes a transformer-based method for zero- and few-shot NER in the biomedical domain. The method is based on transforming the task of multi-class token classification into binary token classification (token contains the searched entity or does not contain the searched entity) and pre-training on a larger amount of datasets and biomedical entities, from where the method can learn semantic relations between the given and potential classes. We have achieved average F1 scores of 35.44% for zero-shot NER, 50.10% for one-shot NER, 69.94% for 10-shot NER, and 79.51% for 100-shot NER on 9 diverse evaluated biomedical entities with PubMedBERT fine-tuned model. The results demonstrate the effectiveness of the proposed method for recognizing new entities with limited examples, with comparable or better results from the state-of-the-art zero- and few-shot NER methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge