Birgitta K. Velthuis
Deep Learning for Automatic Strain Quantification in Arrhythmogenic Right Ventricular Cardiomyopathy
Nov 24, 2023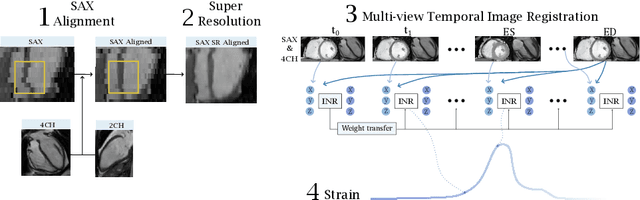
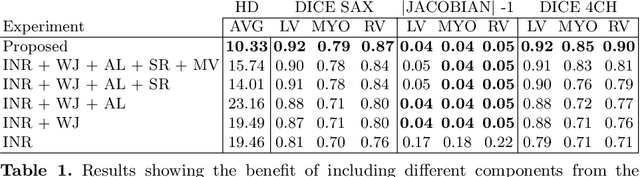
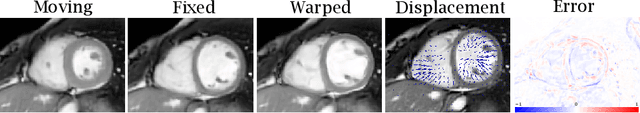
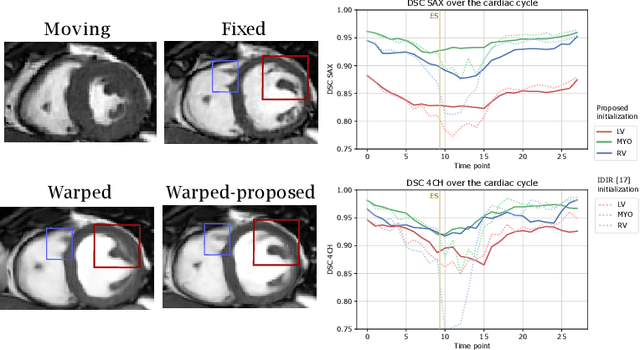
Abstract:Quantification of cardiac motion with cine Cardiac Magnetic Resonance Imaging (CMRI) is an integral part of arrhythmogenic right ventricular cardiomyopathy (ARVC) diagnosis. Yet, the expert evaluation of motion abnormalities with CMRI is a challenging task. To automatically assess cardiac motion, we register CMRIs from different time points of the cardiac cycle using Implicit Neural Representations (INRs) and perform a biomechanically informed regularization inspired by the myocardial incompressibility assumption. To enhance the registration performance, our method first rectifies the inter-slice misalignment inherent to CMRI by performing a rigid registration guided by the long-axis views, and then increases the through-plane resolution using an unsupervised deep learning super-resolution approach. Finally, we propose to synergically combine information from short-axis and 4-chamber long-axis views, along with an initialization to incorporate information from multiple cardiac time points. Thereafter, to quantify cardiac motion, we calculate global and segmental strain over a cardiac cycle and compute the peak strain. The evaluation of the method is performed on a dataset of cine CMRI scans from 47 ARVC patients and 67 controls. Our results show that inter-slice alignment and generation of super-resolved volumes combined with joint analysis of the two cardiac views, notably improves registration performance. Furthermore, the proposed initialization yields more physiologically plausible registrations. The significant differences in the peak strain, discerned between the ARVC patients and healthy controls suggest that automated motion quantification methods may assist in diagnosis and provide further understanding of disease-specific alterations of cardiac motion.
Future Unruptured Intracranial Aneurysm Growth Prediction using Mesh Convolutional Neural Networks
Aug 01, 2022


Abstract:The growth of unruptured intracranial aneurysms (UIAs) is a predictor of rupture. Therefore, for further imaging surveillance and treatment planning, it is important to be able to predict if an UIA is likely to grow based on an initial baseline Time-of-Flight MRA (TOF-MRA). It is known that the size and shape of UIAs are predictors of aneurysm growth and/or rupture. We perform a feasibility study of using a mesh convolutional neural network for future UIA growth prediction from baseline TOF-MRAs. We include 151 TOF-MRAs, with 169 UIAs where 49 UIAs were classified as growing and 120 as stable, based on the clinical definition of growth (>1 mm increase in size in follow-up scan). UIAs were segmented from TOF-MRAs and meshes were automatically generated. We investigate the input of both UIA mesh only and region-of-interest (ROI) meshes including UIA and surrounding parent vessels. We develop a classification model to predict UIAs that will grow or remain stable. The model consisted of a mesh convolutional neural network including additional novel input edge features of shape index and curvedness which describe the surface topology. It was investigated if input edge mid-point co-ordinates influenced the model performance. The model with highest AUC (63.8%) for growth prediction was using UIA meshes with input edge mid-point co-ordinate features (average F1 score = 62.3%, accuracy = 66.9%, sensitivity = 57.3%, specificity = 70.8%). We present a future UIA growth prediction model based on a mesh convolutional neural network with promising results.
Variational Autoencoders with a Structural Similarity Loss in Time of Flight MRAs
Jan 20, 2021



Abstract:Time-of-Flight Magnetic Resonance Angiographs (TOF-MRAs) enable visualization and analysis of cerebral arteries. This analysis may indicate normal variation of the configuration of the cerebrovascular system or vessel abnormalities, such as aneurysms. A model would be useful to represent normal cerebrovascular structure and variabilities in a healthy population and to differentiate from abnormalities. Current anomaly detection using autoencoding convolutional neural networks usually use a voxelwise mean-error for optimization. We propose optimizing a variational-autoencoder (VAE) with structural similarity loss (SSIM) for TOF-MRA reconstruction. A patch-trained 2D fully-convolutional VAE was optimized for TOF-MRA reconstruction by comparing vessel segmentations of original and reconstructed MRAs. The method was trained and tested on two datasets: the IXI dataset, and a subset from the ADAM challenge. Both trained networks were tested on a dataset including subjects with aneurysms. We compared VAE optimization with L2-loss and SSIM-loss. Performance was evaluated between original and reconstructed MRAs using mean square error, mean-SSIM, peak-signal-to-noise-ratio and dice similarity index (DSI) of segmented vessels. The L2-optimized VAE outperforms SSIM, with improved reconstruction metrics and DSIs for both datasets. Optimization using SSIM performed best for visual image quality, but with discrepancy in quantitative reconstruction and vascular segmentation. The larger, more diverse IXI dataset had overall better performance. Reconstruction metrics, including SSIM, were lower for MRAs including aneurysms. A SSIM-optimized VAE improved the visual perceptive image quality of TOF-MRA reconstructions. A L2-optimized VAE performed best for TOF-MRA reconstruction, where the vascular segmentation is important. SSIM is a potential metric for anomaly detection of MRAs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge