Beyrem Khalfaoui
CBIO
ASNI: Adaptive Structured Noise Injection for shallow and deep neural networks
Sep 21, 2019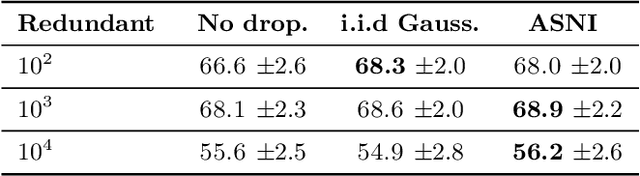
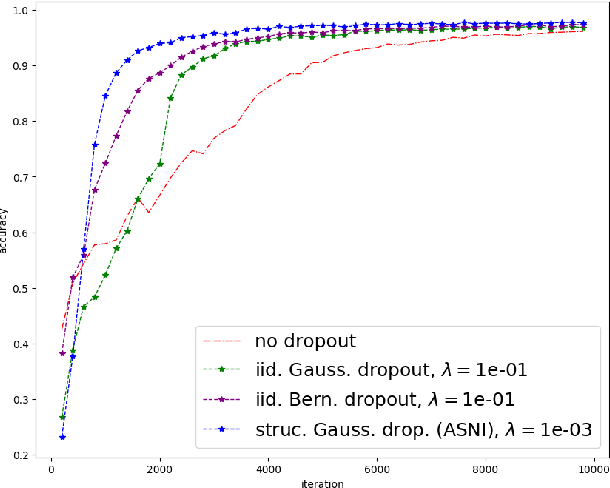
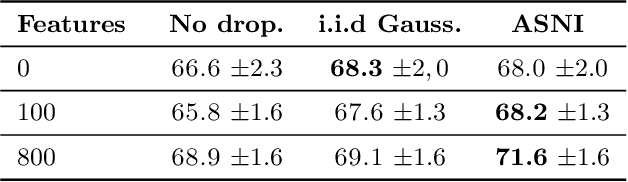
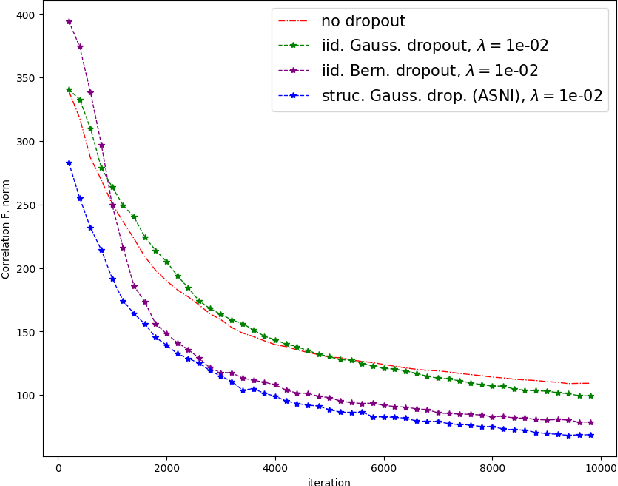
Abstract:Dropout is a regularisation technique in neural network training where unit activations are randomly set to zero with a given probability \emph{independently}. In this work, we propose a generalisation of dropout and other multiplicative noise injection schemes for shallow and deep neural networks, where the random noise applied to different units is not independent but follows a joint distribution that is either fixed or estimated during training. We provide theoretical insights on why such adaptive structured noise injection (ASNI) may be relevant, and empirically confirm that it helps boost the accuracy of simple feedforward and convolutional neural networks, disentangles the hidden layer representations, and leads to sparser representations. Our proposed method is a straightforward modification of the classical dropout and does not require additional computational overhead.
DropLasso: A robust variant of Lasso for single cell RNA-seq data
Feb 26, 2018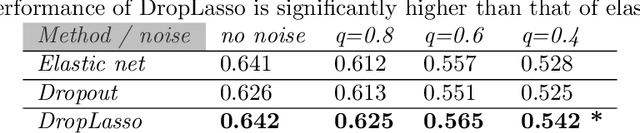
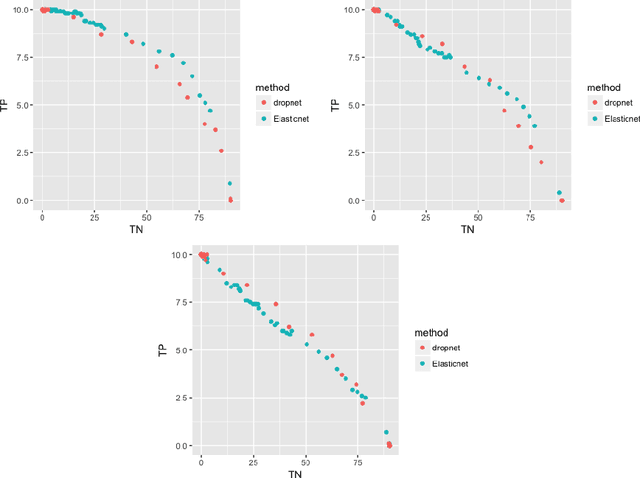
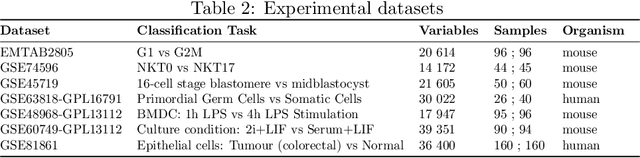
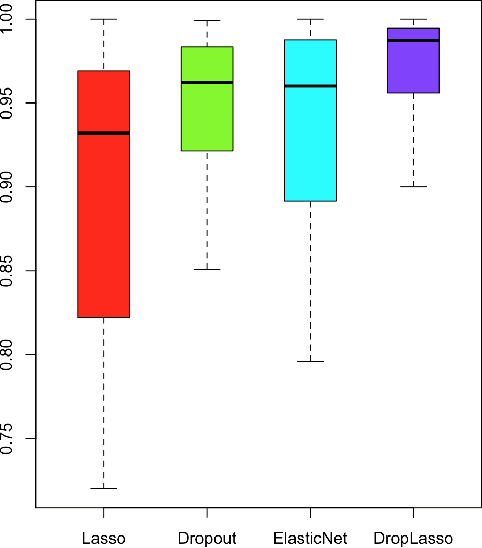
Abstract:Single-cell RNA sequencing (scRNA-seq) is a fast growing approach to measure the genome-wide transcriptome of many individual cells in parallel, but results in noisy data with many dropout events. Existing methods to learn molecular signatures from bulk transcriptomic data may therefore not be adapted to scRNA-seq data, in order to automatically classify individual cells into predefined classes. We propose a new method called DropLasso to learn a molecular signature from scRNA-seq data. DropLasso extends the dropout regularisation technique, popular in neural network training, to esti- mate sparse linear models. It is well adapted to data corrupted by dropout noise, such as scRNA-seq data, and we clarify how it relates to elastic net regularisation. We provide promising results on simulated and real scRNA-seq data, suggesting that DropLasso may be better adapted than standard regularisa- tions to infer molecular signatures from scRNA-seq data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge