Ben Feng
Dynamic Bayesian Network Auxiliary ABC-SMC for Hybrid Model Bayesian Inference to Accelerate Biomanufacturing Process Mechanism Learning and Robust Control
May 09, 2022
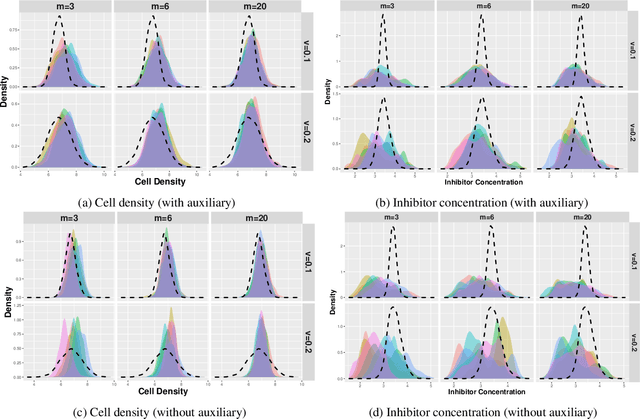

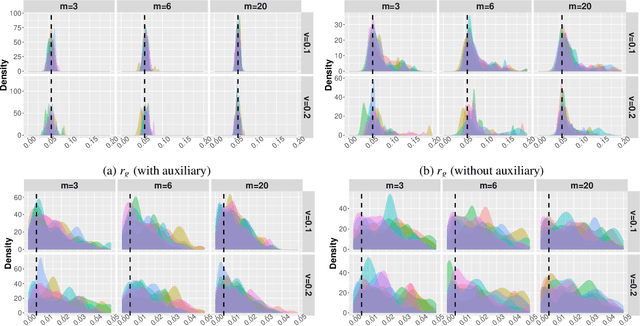
Abstract:Driven by the critical needs of biomanufacturing 4.0, we present a probabilistic knowledge graph hybrid model characterizing complex spatial-temporal causal interdependencies of underlying bioprocessing mechanisms. It can faithfully capture the important properties, including nonlinear reactions, partially observed state, and nonstationary dynamics. Given limited process observations, we derive a posterior distribution quantifying model uncertainty, which can facilitate mechanism learning and support robust process control. To avoid evaluation of intractable likelihood, Approximate Bayesian Computation sampling with Sequential Monte Carlo (ABC-SMC) is developed to approximate the posterior distribution. Given high stochastic and model uncertainties, it is computationally expensive to match process output trajectories. Therefore, we propose a linear Gaussian dynamic Bayesian network (LG-DBN) auxiliary likelihood-based ABC-SMC algorithm. Through matching observed and simulated summary statistics, the proposed approach can dramatically reduce the computation cost and improve the posterior distribution approximation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge