Bastien Chopard
Computer Science Department, Faculty of Science, University of Geneva, Switzerland
SISMIK for brain MRI: Deep-learning-based motion estimation and model-based motion correction in k-space
Dec 20, 2023Abstract:MRI, a widespread non-invasive medical imaging modality, is highly sensitive to patient motion. Despite many attempts over the years, motion correction remains a difficult problem and there is no general method applicable to all situations. We propose a retrospective method for motion quantification and correction to tackle the problem of in-plane rigid-body motion, apt for classical 2D Spin-Echo scans of the brain, which are regularly used in clinical practice. Due to the sequential acquisition of k-space, motion artifacts are well localized. The method leverages the power of deep neural networks to estimate motion parameters in k-space and uses a model-based approach to restore degraded images to avoid ''hallucinations''. Notable advantages are its ability to estimate motion occurring in high spatial frequencies without the need of a motion-free reference. The proposed method operates on the whole k-space dynamic range and is moderately affected by the lower SNR of higher harmonics. As a proof of concept, we provide models trained using supervised learning on 600k motion simulations based on motion-free scans of 43 different subjects. Generalization performance was tested with simulations as well as in-vivo. Qualitative and quantitative evaluations are presented for motion parameter estimations and image reconstruction. Experimental results show that our approach is able to obtain good generalization performance on simulated data and in-vivo acquisitions.
Interpretable pathological test for Cardio-vascular disease: Approximate Bayesian computation with distance learning
Oct 13, 2020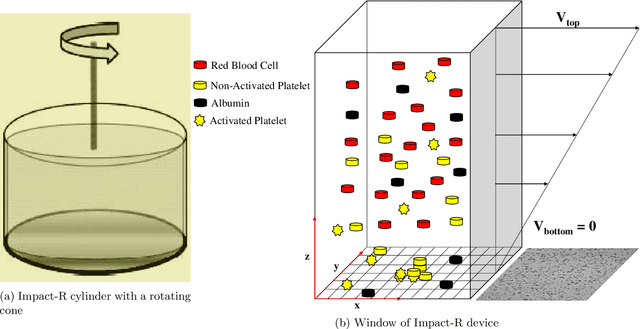
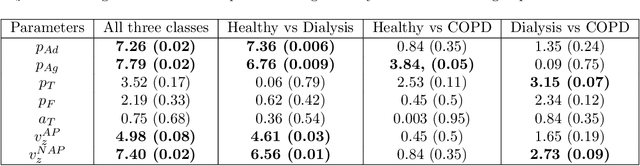
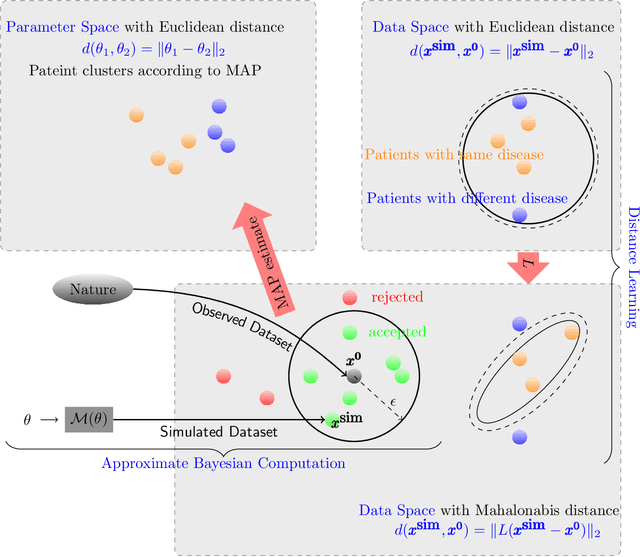

Abstract:Cardio/cerebrovascular diseases (CVD) have become one of the major health issue in our societies. But recent studies show that the present clinical tests to detect CVD are ineffectual as they do not consider different stages of platelet activation or the molecular dynamics involved in platelet interactions and are incapable to consider inter-individual variability. Here we propose a stochastic platelet deposition model and an inferential scheme for uncertainty quantification of these parameters using Approximate Bayesian Computation and distance learning. Finally we show that our methodology can learn biologically meaningful parameters, which are the specific dysfunctioning parameters in each type of patients, from data collected from healthy volunteers and patients. This work opens up an unprecedented opportunity of personalized pathological test for CVD detection and medical treatment. Also our proposed methodology can be used to other fields of science where we would need machine learning tools to be interpretable.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge