Barbara Koroušić Seljak
FoodSEM: Large Language Model Specialized in Food Named-Entity Linking
Sep 26, 2025Abstract:This paper introduces FoodSEM, a state-of-the-art fine-tuned open-source large language model (LLM) for named-entity linking (NEL) to food-related ontologies. To the best of our knowledge, food NEL is a task that cannot be accurately solved by state-of-the-art general-purpose (large) language models or custom domain-specific models/systems. Through an instruction-response (IR) scenario, FoodSEM links food-related entities mentioned in a text to several ontologies, including FoodOn, SNOMED-CT, and the Hansard taxonomy. The FoodSEM model achieves state-of-the-art performance compared to related models/systems, with F1 scores even reaching 98% on some ontologies and datasets. The presented comparative analyses against zero-shot, one-shot, and few-shot LLM prompting baselines further highlight FoodSEM's superior performance over its non-fine-tuned version. By making FoodSEM and its related resources publicly available, the main contributions of this article include (1) publishing a food-annotated corpora into an IR format suitable for LLM fine-tuning/evaluation, (2) publishing a robust model to advance the semantic understanding of text in the food domain, and (3) providing a strong baseline on food NEL for future benchmarking.
Predefined domain specific embeddings of food concepts and recipes: A case study on heterogeneous recipe datasets
Feb 02, 2023Abstract:Although recipe data are very easy to come by nowadays, it is really hard to find a complete recipe dataset - with a list of ingredients, nutrient values per ingredient, and per recipe, allergens, etc. Recipe datasets are usually collected from social media websites where users post and publish recipes. Usually written with little to no structure, using both standardized and non-standardized units of measurement. We collect six different recipe datasets, publicly available, in different formats, and some including data in different languages. Bringing all of these datasets to the needed format for applying a machine learning (ML) pipeline for nutrient prediction [1], [2], includes data normalization using dictionary-based named entity recognition (NER), rule-based NER, as well as conversions using external domain-specific resources. From the list of ingredients, domain-specific embeddings are created using the same embedding space for all recipes - one ingredient dataset is generated. The result from this normalization process is two corpora - one with predefined ingredient embeddings and one with predefined recipe embeddings. On all six recipe datasets, the ML pipeline is evaluated. The results from this use case also confirm that the embeddings merged using the domain heuristic yield better results than the baselines.
FoodChem: A food-chemical relation extraction model
Oct 08, 2021

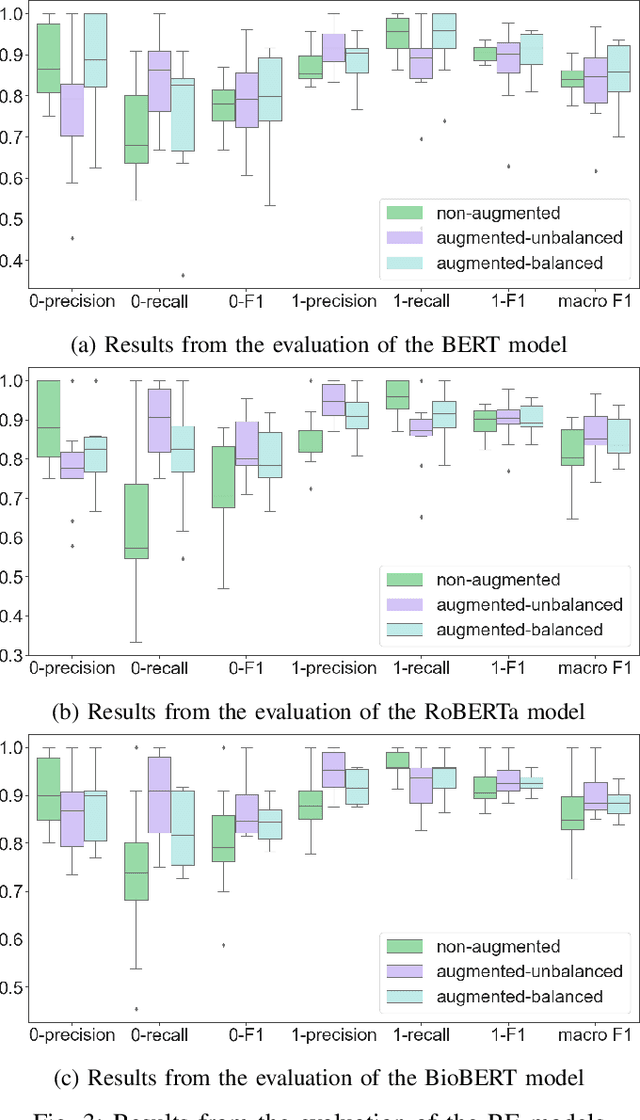
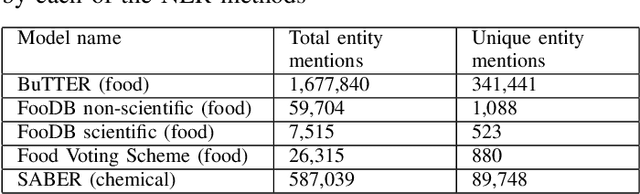
Abstract:In this paper, we present FoodChem, a new Relation Extraction (RE) model for identifying chemicals present in the composition of food entities, based on textual information provided in biomedical peer-reviewed scientific literature. The RE task is treated as a binary classification problem, aimed at identifying whether the contains relation exists between a food-chemical entity pair. This is accomplished by fine-tuning BERT, BioBERT and RoBERTa transformer models. For evaluation purposes, a novel dataset with annotated contains relations in food-chemical entity pairs is generated, in a golden and silver version. The models are integrated into a voting scheme in order to produce the silver version of the dataset which we use for augmenting the individual models, while the manually annotated golden version is used for their evaluation. Out of the three evaluated models, the BioBERT model achieves the best results, with a macro averaged F1 score of 0.902 in the unbalanced augmentation setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge