Asmaa Haja
Self-Supervised Versus Supervised Training for Segmentation of Organoid Images
Nov 19, 2023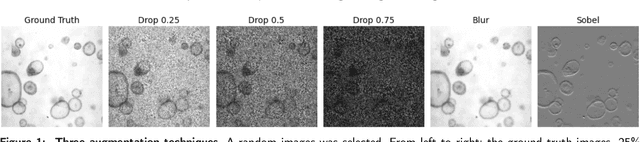
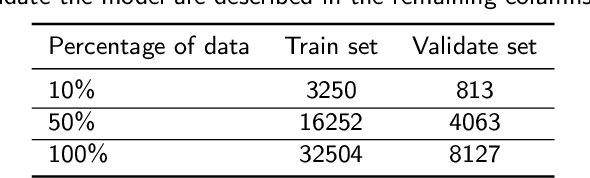
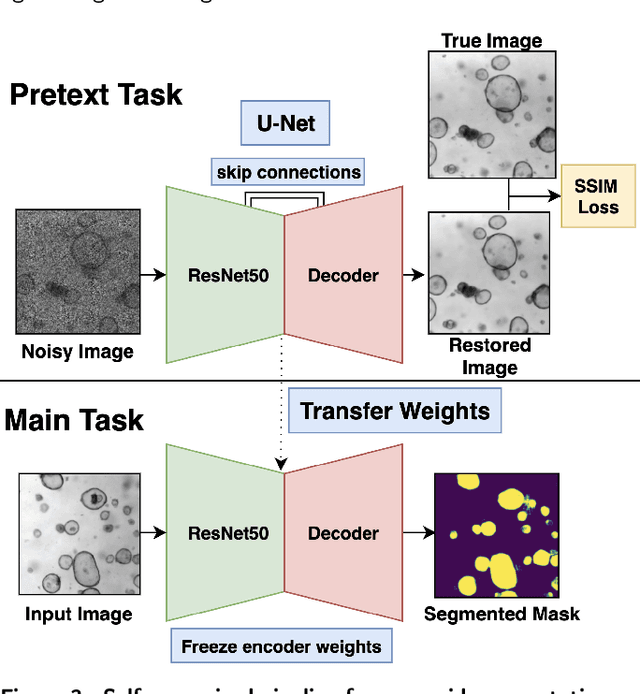
Abstract:The process of annotating relevant data in the field of digital microscopy can be both time-consuming and especially expensive due to the required technical skills and human-expert knowledge. Consequently, large amounts of microscopic image data sets remain unlabeled, preventing their effective exploitation using deep-learning algorithms. In recent years it has been shown that a lot of relevant information can be drawn from unlabeled data. Self-supervised learning (SSL) is a promising solution based on learning intrinsic features under a pretext task that is similar to the main task without requiring labels. The trained result is transferred to the main task - image segmentation in our case. A ResNet50 U-Net was first trained to restore images of liver progenitor organoids from augmented images using the Structural Similarity Index Metric (SSIM), alone, and using SSIM combined with L1 loss. Both the encoder and decoder were trained in tandem. The weights were transferred to another U-Net model designed for segmentation with frozen encoder weights, using Binary Cross Entropy, Dice, and Intersection over Union (IoU) losses. For comparison, we used the same U-Net architecture to train two supervised models, one utilizing the ResNet50 encoder as well as a simple CNN. Results showed that self-supervised learning models using a 25\% pixel drop or image blurring augmentation performed better than the other augmentation techniques using the IoU loss. When trained on only 114 images for the main task, the self-supervised learning approach outperforms the supervised method achieving an F1-score of 0.85, with higher stability, in contrast to an F1=0.78 scored by the supervised method. Furthermore, when trained with larger data sets (1,000 images), self-supervised learning is still able to perform better, achieving an F1-score of 0.92, contrasting to a score of 0.85 for the supervised method.
A fully automated end-to-end process for fluorescence microscopy images of yeast cells: From segmentation to detection and classification
Apr 06, 2021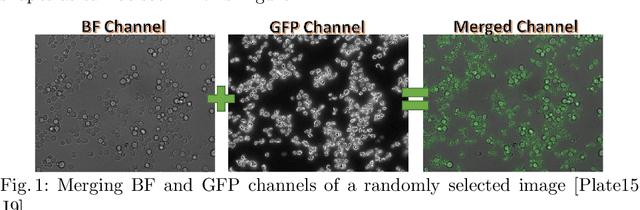

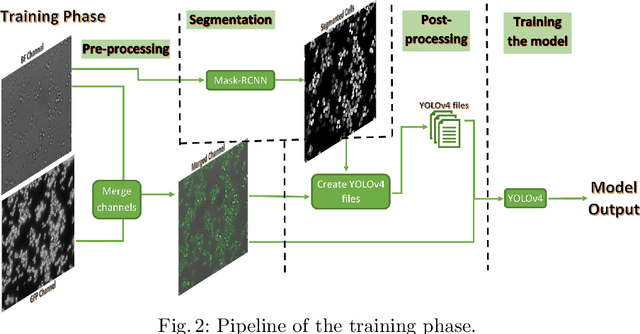
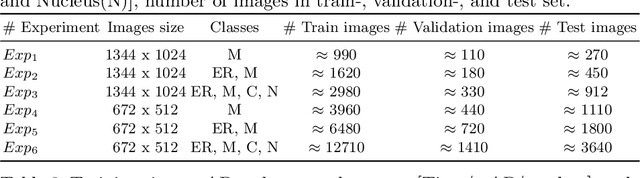
Abstract:In recent years, an enormous amount of fluorescence microscopy images were collected in high-throughput lab settings. Analyzing and extracting relevant information from all images in a short time is almost impossible. Detecting tiny individual cell compartments is one of many challenges faced by biologists. This paper aims at solving this problem by building an end-to-end process that employs methods from the deep learning field to automatically segment, detect and classify cell compartments of fluorescence microscopy images of yeast cells. With this intention we used Mask R-CNN to automatically segment and label a large amount of yeast cell data, and YOLOv4 to automatically detect and classify individual yeast cell compartments from these images. This fully automated end-to-end process is intended to be integrated into an interactive e-Science server in the PerICo1 project, which can be used by biologists with minimized human effort in training and operation to complete their various classification tasks. In addition, we evaluated the detection and classification performance of state-of-the-art YOLOv4 on data from the NOP1pr-GFP-SWAT yeast-cell data library. Experimental results show that by dividing original images into 4 quadrants YOLOv4 outputs good detection and classification results with an F1-score of 98% in terms of accuracy and speed, which is optimally suited for the native resolution of the microscope and current GPU memory sizes. Although the application domain is optical microscopy in yeast cells, the method is also applicable to multiple-cell images in medical applications
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge