Ashesh Ashesh
indiSplit: Bringing Severity Cognizance to Image Decomposition in Fluorescence Microscopy
Mar 29, 2025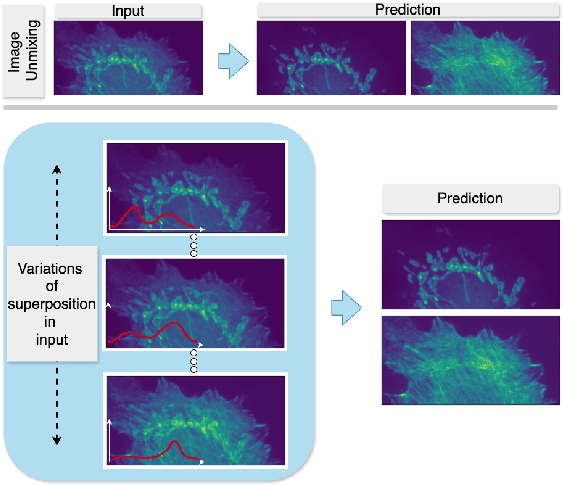
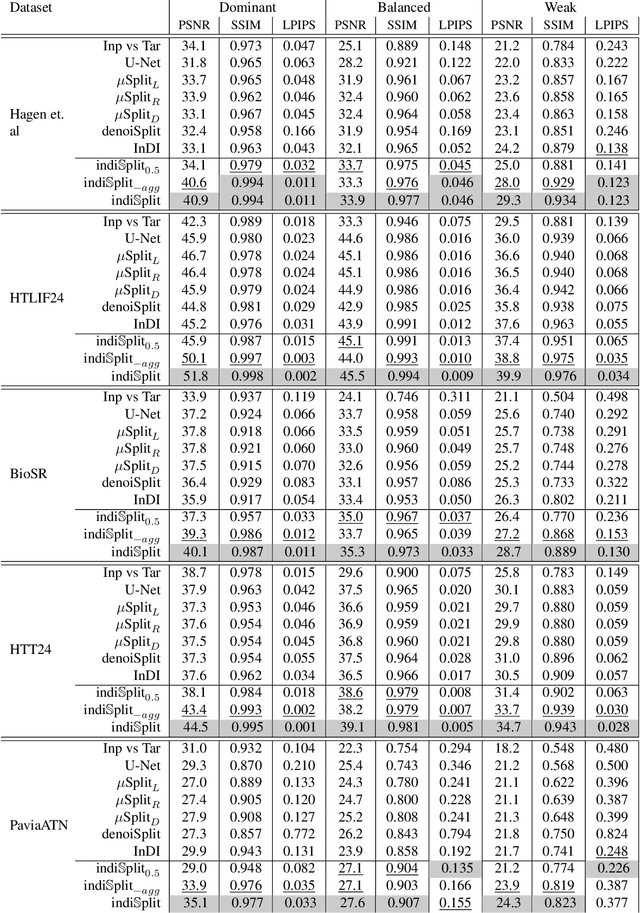
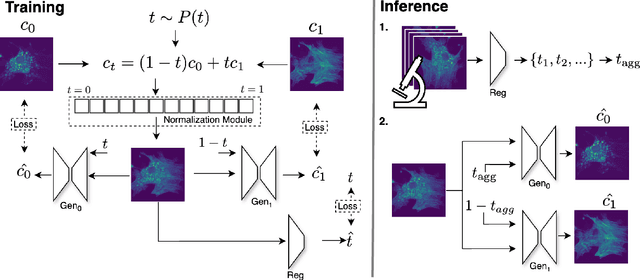
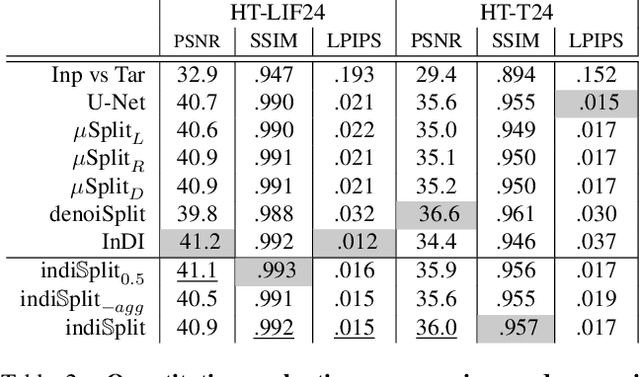
Abstract:Fluorescence microscopy, while being a key driver for progress in the life sciences, is also subject to technical limitations. To overcome them, computational multiplexing techniques have recently been proposed, which allow multiple cellular structures to be captured in a single image and later be unmixed. Existing image decomposition methods are trained on a set of superimposed input images and the respective unmixed target images. It is critical to note that the relative strength (mixing ratio) of the superimposed images for a given input is a priori unknown. However, existing methods are trained on a fixed intensity ratio of superimposed inputs, making them not cognizant to the range of relative intensities that can occur in fluorescence microscopy. In this work, we propose a novel method called indiSplit that is cognizant of the severity of the above mentioned mixing ratio. Our idea is based on InDI, a popular iterative method for image restoration, and an ideal starting point to embrace the unknown mixing ratio in any given input. We introduce (i) a suitably trained regressor network that predicts the degradation level (mixing asymmetry) of a given input image and (ii) a degradation-specific normalization module, enabling degradation-aware inference across all mixing ratios. We show that this method solves two relevant tasks in fluorescence microscopy, namely image splitting and bleedthrough removal, and empirically demonstrate the applicability of indiSplit on $5$ public datasets. We will release all sources under a permissive license.
MicroSSIM: Improved Structural Similarity for Comparing Microscopy Data
Aug 16, 2024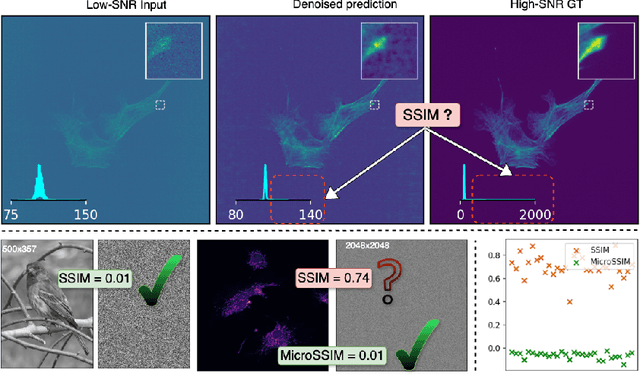
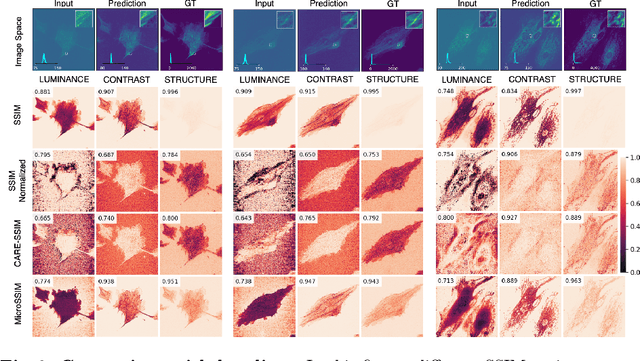
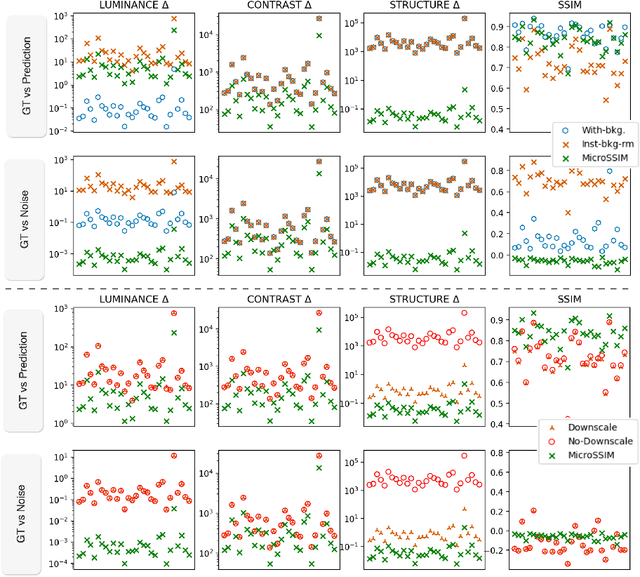
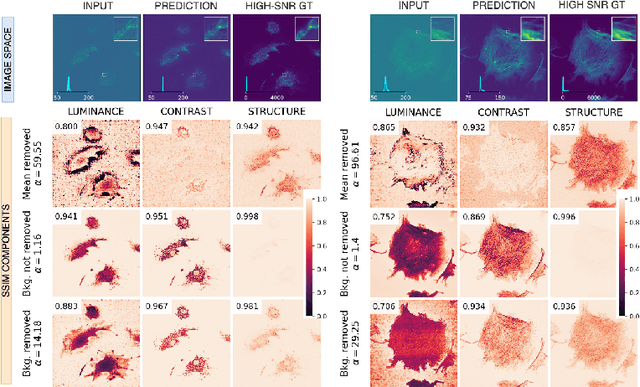
Abstract:Microscopy is routinely used to image biological structures of interest. Due to imaging constraints, acquired images are typically low-SNR and contain noise. Over the last few years, regression-based tasks like unsupervised denoising and splitting have found utility in working with such noisy micrographs. For evaluation, Structural Similarity (SSIM) is one of the most popular measures used in the field. For such tasks, the best evaluation would be when both low-SNR noisy images and corresponding high-SNR clean images are obtained directly from a microscope. However, due to the following three peculiar properties of the microscopy data, we observe that SSIM is not well suited to this data regime: (a) high-SNR micrographs have higher intensity pixels as compared to low SNR micrographs, (b) high-SNR micrographs have higher intensity pixels than found in natural images, images for which SSIM was developed, and (c) a digitally configurable offset is added by the detector present inside the microscope. We show that SSIM components behave unexpectedly when the prediction generated from low-SNR input is compared with the corresponding high-SNR data. We explain this behavior by introducing the phenomenon of saturation, where the value of SSIM components becomes less sensitive to (dis)similarity between the images. We introduce microSSIM, a variant of SSIM, which overcomes the above-discussed issues. We justify the soundness and utility of microSSIM using theoretical and empirical arguments and show the utility of microSSIM on two tasks: unsupervised denoising and joint image splitting with unsupervised denoising. Since our formulation can be applied to a broad family of SSIM-based measures, we also introduce MicroMS3IM, a microscopy-specific variation of MS-SSIM. The source code and python package is available at https://github.com/juglab/MicroSSIM.
denoiSplit: a method for joint image splitting and unsupervised denoising
Mar 25, 2024Abstract:In this work we present denoiSplit, a method to tackle a new analysis task, i.e. the challenge of joint semantic image splitting and unsupervised denoising. This dual approach has important applications in fluorescence microscopy, where semantic image splitting has important applications but noise does generally hinder the downstream analysis of image content. Image splitting involves dissecting an image into its distinguishable semantic structures. We show that the current state-of-the-art method for this task struggles in the presence of image noise, inadvertently also distributing the noise across the predicted outputs. The method we present here can deal with image noise by integrating an unsupervised denoising sub-task. This integration results in improved semantic image unmixing, even in the presence of notable and realistic levels of imaging noise. A key innovation in denoiSplit is the use of specifically formulated noise models and the suitable adjustment of KL-divergence loss for the high-dimensional hierarchical latent space we are training. We showcase the performance of denoiSplit across 4 tasks on real-world microscopy images. Additionally, we perform qualitative and quantitative evaluations and compare results to existing benchmarks, demonstrating the effectiveness of using denoiSplit: a single Variational Splitting Encoder-Decoder (VSE) Network using two suitable noise models to jointly perform semantic splitting and denoising.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge