MicroSSIM: Improved Structural Similarity for Comparing Microscopy Data
Paper and Code
Aug 16, 2024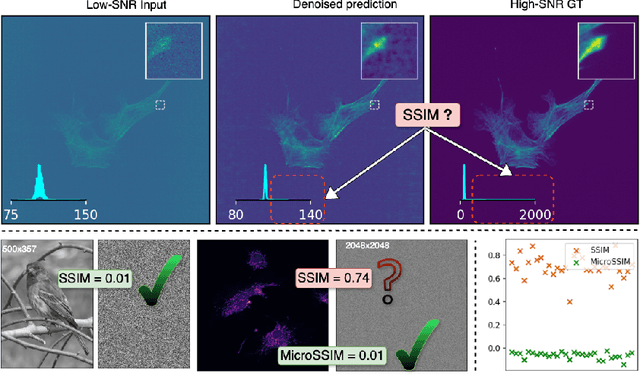
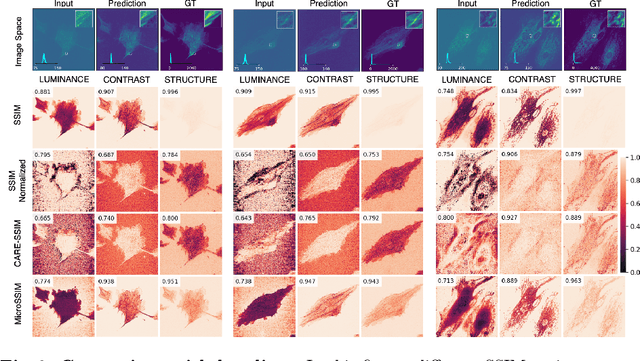
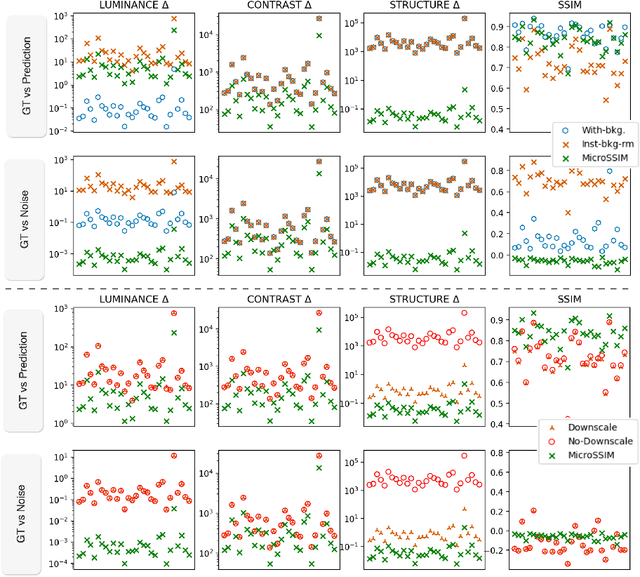
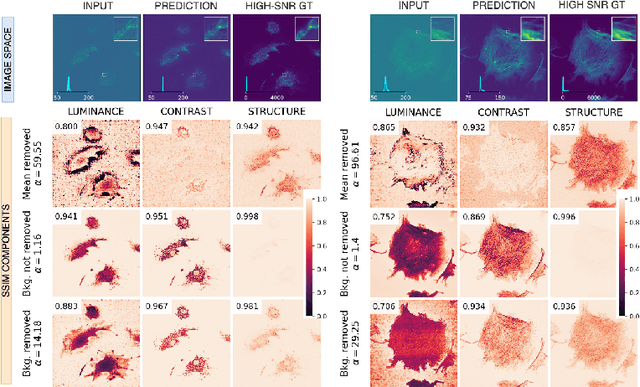
Microscopy is routinely used to image biological structures of interest. Due to imaging constraints, acquired images are typically low-SNR and contain noise. Over the last few years, regression-based tasks like unsupervised denoising and splitting have found utility in working with such noisy micrographs. For evaluation, Structural Similarity (SSIM) is one of the most popular measures used in the field. For such tasks, the best evaluation would be when both low-SNR noisy images and corresponding high-SNR clean images are obtained directly from a microscope. However, due to the following three peculiar properties of the microscopy data, we observe that SSIM is not well suited to this data regime: (a) high-SNR micrographs have higher intensity pixels as compared to low SNR micrographs, (b) high-SNR micrographs have higher intensity pixels than found in natural images, images for which SSIM was developed, and (c) a digitally configurable offset is added by the detector present inside the microscope. We show that SSIM components behave unexpectedly when the prediction generated from low-SNR input is compared with the corresponding high-SNR data. We explain this behavior by introducing the phenomenon of saturation, where the value of SSIM components becomes less sensitive to (dis)similarity between the images. We introduce microSSIM, a variant of SSIM, which overcomes the above-discussed issues. We justify the soundness and utility of microSSIM using theoretical and empirical arguments and show the utility of microSSIM on two tasks: unsupervised denoising and joint image splitting with unsupervised denoising. Since our formulation can be applied to a broad family of SSIM-based measures, we also introduce MicroMS3IM, a microscopy-specific variation of MS-SSIM. The source code and python package is available at https://github.com/juglab/MicroSSIM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge