Arunkumar Govindarajan
Identification of Hemorrhage and Infarct Lesions on Brain CT Images using Deep Learning
Jul 10, 2023Abstract:Head Non-contrast computed tomography (NCCT) scan remain the preferred primary imaging modality due to their widespread availability and speed. However, the current standard for manual annotations of abnormal brain tissue on head NCCT scans involves significant disadvantages like lack of cutoff standardization and degeneration identification. The recent advancement of deep learning-based computer-aided diagnostic (CAD) models in the multidisciplinary domain has created vast opportunities in neurological medical imaging. Significant literature has been published earlier in the automated identification of brain tissue on different imaging modalities. However, determining Intracranial hemorrhage (ICH) and infarct can be challenging due to image texture, volume size, and scan quality variability. This retrospective validation study evaluated a DL-based algorithm identifying ICH and infarct from head-NCCT scans. The head-NCCT scans dataset was collected consecutively from multiple diagnostic imaging centers across India. The study exhibits the potential and limitations of such DL-based software for introduction in routine workflow in extensive healthcare facilities.
Towards A Device-Independent Deep Learning Approach for the Automated Segmentation of Sonographic Fetal Brain Structures: A Multi-Center and Multi-Device Validation
Feb 28, 2022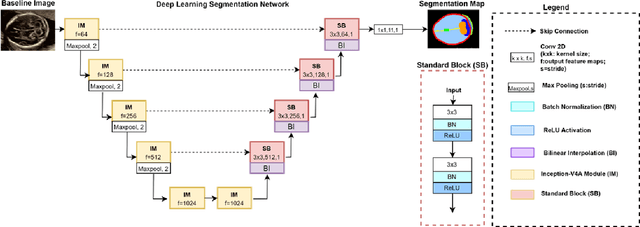
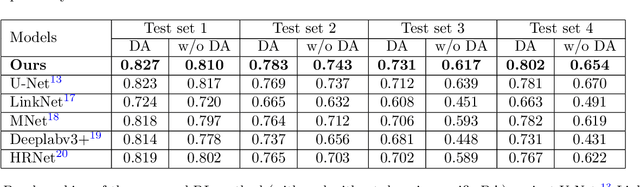
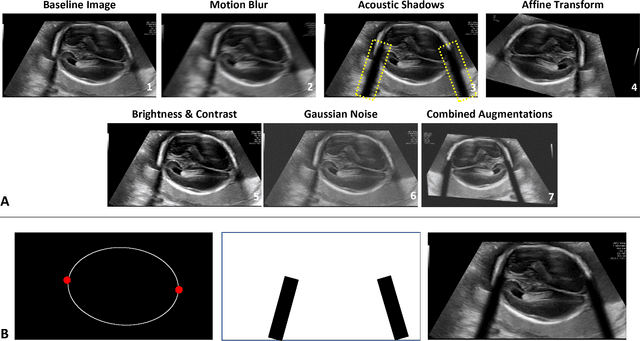
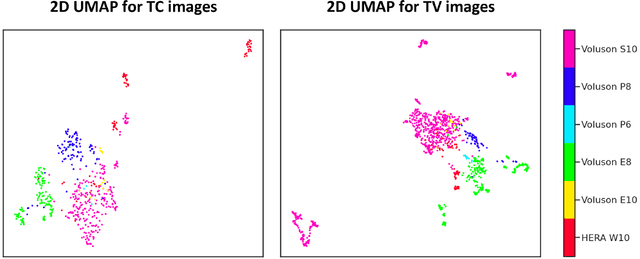
Abstract:Quality assessment of prenatal ultrasonography is essential for the screening of fetal central nervous system (CNS) anomalies. The interpretation of fetal brain structures is highly subjective, expertise-driven, and requires years of training experience, limiting quality prenatal care for all pregnant mothers. With recent advancement in Artificial Intelligence (AI), specifically deep learning (DL), assistance in precise anatomy identification through semantic segmentation essential for the reliable assessment of growth and neurodevelopment, and detection of structural abnormalities have been proposed. However, existing works only identify certain structures (e.g., cavum septum pellucidum, lateral ventricles, cerebellum) from either of the axial views (transventricular, transcerebellar), limiting the scope for a thorough anatomical assessment as per practice guidelines necessary for the screening of CNS anomalies. Further, existing works do not analyze the generalizability of these DL algorithms across images from multiple ultrasound devices and centers, thus, limiting their real-world clinical impact. In this study, we propose a DL based segmentation framework for the automated segmentation of 10 key fetal brain structures from 2 axial planes from fetal brain USG images (2D). We developed a custom U-Net variant that uses inceptionv4 block as a feature extractor and leverages custom domain-specific data augmentation. Quantitatively, the mean (10 structures; test sets 1/2/3/4) Dice-coefficients were: 0.827, 0.802, 0.731, 0.783. Irrespective of the USG device/center, the DL segmentations were qualitatively comparable to their manual segmentations. The proposed DL system offered a promising and generalizable performance (multi-centers, multi-device) and also presents evidence in support of device-induced variation in image quality (a challenge to generalizibility) by using UMAP analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge