Arun Palla
Generalizing Supervised Deep Learning MRI Reconstruction to Multiple and Unseen Contrasts using Meta-Learning Hypernetworks
Jul 13, 2023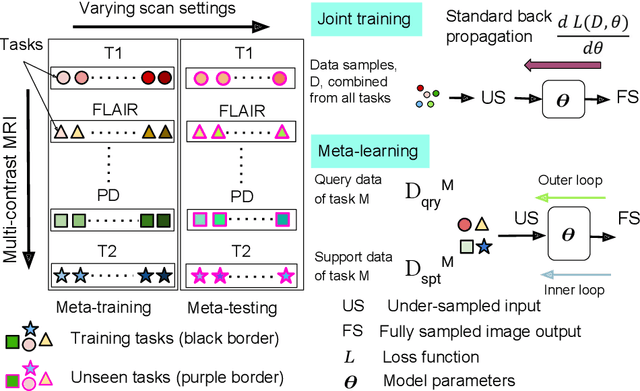
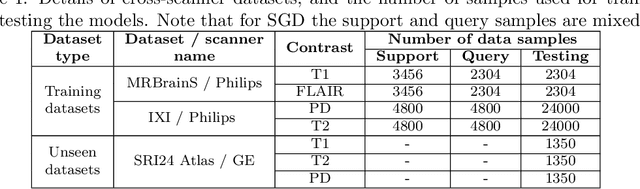
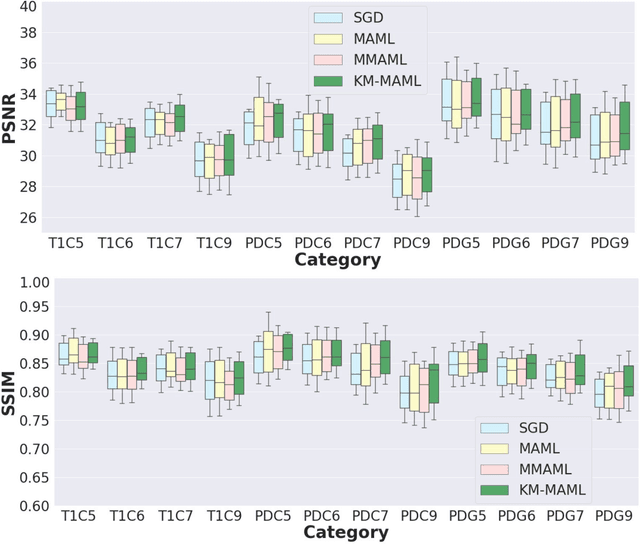
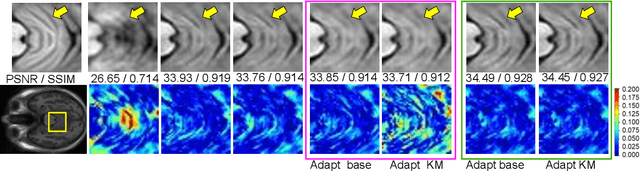
Abstract:Meta-learning has recently been an emerging data-efficient learning technique for various medical imaging operations and has helped advance contemporary deep learning models. Furthermore, meta-learning enhances the knowledge generalization of the imaging tasks by learning both shared and discriminative weights for various configurations of imaging tasks. However, existing meta-learning models attempt to learn a single set of weight initializations of a neural network that might be restrictive for multimodal data. This work aims to develop a multimodal meta-learning model for image reconstruction, which augments meta-learning with evolutionary capabilities to encompass diverse acquisition settings of multimodal data. Our proposed model called KM-MAML (Kernel Modulation-based Multimodal Meta-Learning), has hypernetworks that evolve to generate mode-specific weights. These weights provide the mode-specific inductive bias for multiple modes by re-calibrating each kernel of the base network for image reconstruction via a low-rank kernel modulation operation. We incorporate gradient-based meta-learning (GBML) in the contextual space to update the weights of the hypernetworks for different modes. The hypernetworks and the reconstruction network in the GBML setting provide discriminative mode-specific features and low-level image features, respectively. Experiments on multi-contrast MRI reconstruction show that our model, (i) exhibits superior reconstruction performance over joint training, other meta-learning methods, and context-specific MRI reconstruction methods, and (ii) better adaptation capabilities with improvement margins of 0.5 dB in PSNR and 0.01 in SSIM. Besides, a representation analysis with U-Net shows that kernel modulation infuses 80% of mode-specific representation changes in the high-resolution layers. Our source code is available at https://github.com/sriprabhar/KM-MAML/.
Generalizable Deep Learning Method for Suppressing Unseen and Multiple MRI Artifacts Using Meta-learning
Apr 13, 2023



Abstract:Magnetic Resonance (MR) images suffer from various types of artifacts due to motion, spatial resolution, and under-sampling. Conventional deep learning methods deal with removing a specific type of artifact, leading to separately trained models for each artifact type that lack the shared knowledge generalizable across artifacts. Moreover, training a model for each type and amount of artifact is a tedious process that consumes more training time and storage of models. On the other hand, the shared knowledge learned by jointly training the model on multiple artifacts might be inadequate to generalize under deviations in the types and amounts of artifacts. Model-agnostic meta-learning (MAML), a nested bi-level optimization framework is a promising technique to learn common knowledge across artifacts in the outer level of optimization, and artifact-specific restoration in the inner level. We propose curriculum-MAML (CMAML), a learning process that integrates MAML with curriculum learning to impart the knowledge of variable artifact complexity to adaptively learn restoration of multiple artifacts during training. Comparative studies against Stochastic Gradient Descent and MAML, using two cardiac datasets reveal that CMAML exhibits (i) better generalization with improved PSNR for 83% of unseen types and amounts of artifacts and improved SSIM in all cases, and (ii) better artifact suppression in 4 out of 5 cases of composite artifacts (scans with multiple artifacts).
A Study of Representational Properties of Unsupervised Anomaly Detection in Brain MRI
Nov 28, 2022



Abstract:Anomaly detection in MRI is of high clinical value in imaging and diagnosis. Unsupervised methods for anomaly detection provide interesting formulations based on reconstruction or latent embedding, offering a way to observe properties related to factorization. We study four existing modeling methods, and report our empirical observations using simple data science tools, to seek outcomes from the perspective of factorization as it would be most relevant to the task of unsupervised anomaly detection, considering the case of brain structural MRI. Our study indicates that anomaly detection algorithms that exhibit factorization related properties are well capacitated with delineatory capabilities to distinguish between normal and anomaly data. We have validated our observations in multiple anomaly and normal datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge