Arthur Spencer
The Virtual Patch Clamp: Imputing C. elegans Membrane Potentials from Calcium Imaging
Jul 24, 2019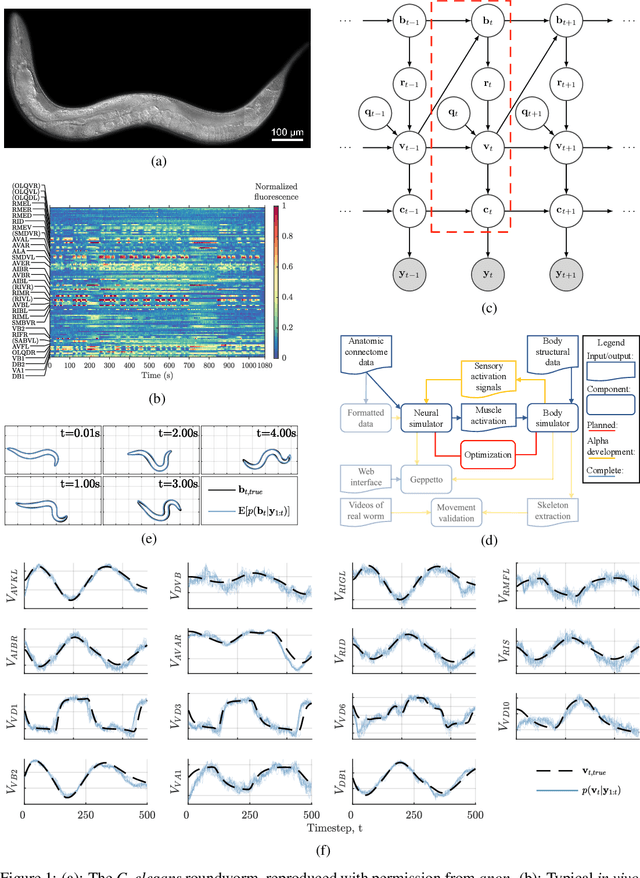
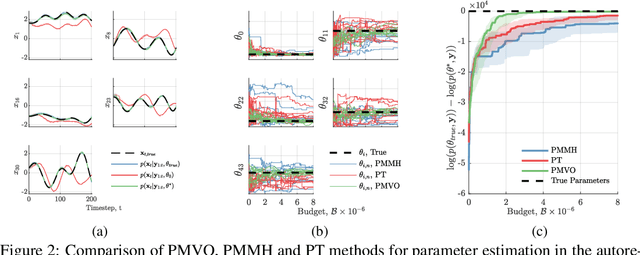
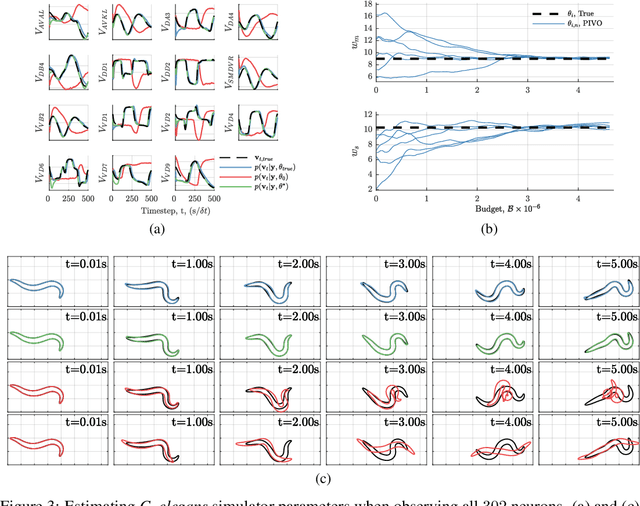
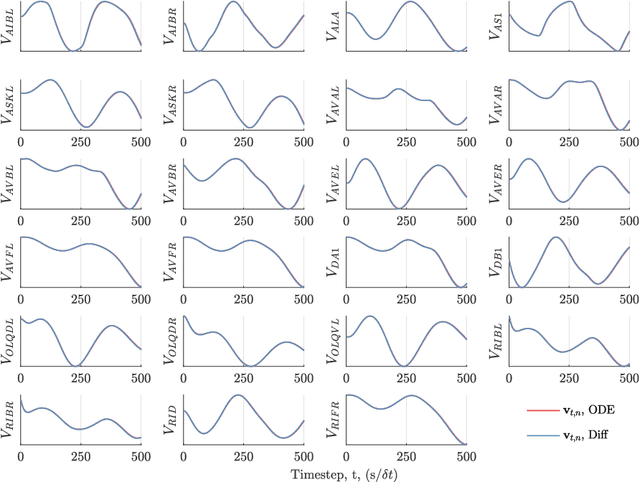
Abstract:We develop a stochastic whole-brain and body simulator of the nematode roundworm Caenorhabditis elegans (C. elegans) and show that it is sufficiently regularizing to allow imputation of latent membrane potentials from partial calcium fluorescence imaging observations. This is the first attempt we know of to "complete the circle," where an anatomically grounded whole-connectome simulator is used to impute a time-varying "brain" state at single-cell fidelity from covariates that are measurable in practice. The sequential Monte Carlo (SMC) method we employ not only enables imputation of said latent states but also presents a strategy for learning simulator parameters via variational optimization of the noisy model evidence approximation provided by SMC. Our imputation and parameter estimation experiments were conducted on distributed systems using novel implementations of the aforementioned techniques applied to synthetic data of dimension and type representative of that which are measured in laboratories currently.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge