Arthur Boschet
Unpaired Modality Translation for Pseudo Labeling of Histology Images
Dec 03, 2024Abstract:The segmentation of histological images is critical for various biomedical applications, yet the lack of annotated data presents a significant challenge. We propose a microscopy pseudo labeling pipeline utilizing unsupervised image translation to address this issue. Our method generates pseudo labels by translating between labeled and unlabeled domains without requiring prior annotation in the target domain. We evaluate two pseudo labeling strategies across three image domains increasingly dissimilar from the labeled data, demonstrating their effectiveness. Notably, our method achieves a mean Dice score of $0.736 \pm 0.005$ on a SEM dataset using the tutoring path, which involves training a segmentation model on synthetic data created by translating the labeled dataset (TEM) to the target modality (SEM). This approach aims to accelerate the annotation process by providing high-quality pseudo labels as a starting point for manual refinement.
Multi-Domain Data Aggregation for Axon and Myelin Segmentation in Histology Images
Sep 17, 2024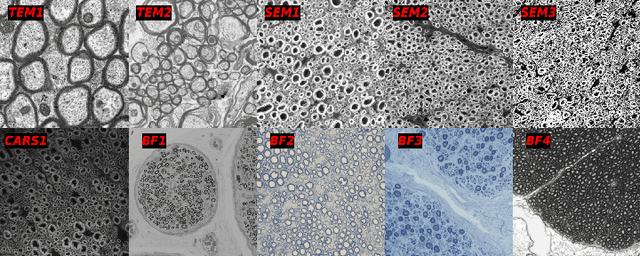
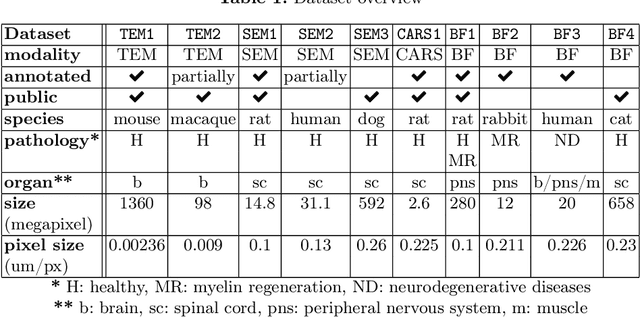
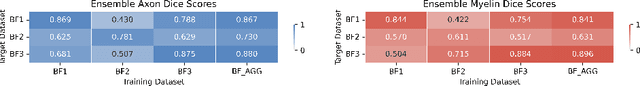
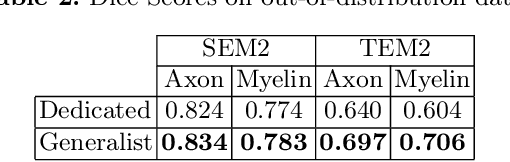
Abstract:Quantifying axon and myelin properties (e.g., axon diameter, myelin thickness, g-ratio) in histology images can provide useful information about microstructural changes caused by neurodegenerative diseases. Automatic tissue segmentation is an important tool for these datasets, as a single stained section can contain up to thousands of axons. Advances in deep learning have made this task quick and reliable with minimal overhead, but a deep learning model trained by one research group will hardly ever be usable by other groups due to differences in their histology training data. This is partly due to subject diversity (different body parts, species, genetics, pathologies) and also to the range of modern microscopy imaging techniques resulting in a wide variability of image features (i.e., contrast, resolution). There is a pressing need to make AI accessible to neuroscience researchers to facilitate and accelerate their workflow, but publicly available models are scarce and poorly maintained. Our approach is to aggregate data from multiple imaging modalities (bright field, electron microscopy, Raman spectroscopy) and species (mouse, rat, rabbit, human), to create an open-source, durable tool for axon and myelin segmentation. Our generalist model makes it easier for researchers to process their data and can be fine-tuned for better performance on specific domains. We study the benefits of different aggregation schemes. This multi-domain segmentation model performs better than single-modality dedicated learners (p=0.03077), generalizes better on out-of-distribution data and is easier to use and maintain. Importantly, we package the segmentation tool into a well-maintained open-source software ecosystem (see https://github.com/axondeepseg/axondeepseg).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge