Aparajita Ojha
A Hybrid Convolutional Neural Network with Meta Feature Learning for Abnormality Detection in Wireless Capsule Endoscopy Images
Jul 20, 2022



Abstract:Wireless Capsule Endoscopy is one of the most advanced non-invasive methods for the examination of gastrointestinal tracts. An intelligent computer-aided diagnostic system for detecting gastrointestinal abnormalities like polyp, bleeding, inflammation, etc. is highly exigent in wireless capsule endoscopy image analysis. Abnormalities greatly differ in their shape, size, color, and texture, and some appear to be visually similar to normal regions. This poses a challenge in designing a binary classifier due to intra-class variations. In this study, a hybrid convolutional neural network is proposed for abnormality detection that extracts a rich pool of meaningful features from wireless capsule endoscopy images using a variety of convolution operations. It consists of three parallel convolutional neural networks, each with a distinctive feature learning capability. The first network utilizes depthwise separable convolution, while the second employs cosine normalized convolution operation. A novel meta-feature extraction mechanism is introduced in the third network, to extract patterns from the statistical information drawn over the features generated from the first and second networks and its own previous layer. The network trio effectively handles intra-class variance and efficiently detects gastrointestinal abnormalities. The proposed hybrid convolutional neural network model is trained and tested on two widely used publicly available datasets. The test results demonstrate that the proposed model outperforms six state-of-the-art methods with 97\% and 98\% classification accuracy on KID and Kvasir-Capsule datasets respectively. Cross dataset evaluation results also demonstrate the generalization performance of the proposed model.
Explainable vision transformer enabled convolutional neural network for plant disease identification: PlantXViT
Jul 16, 2022
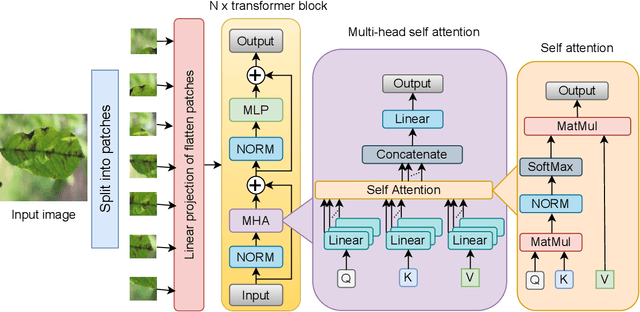


Abstract:Plant diseases are the primary cause of crop losses globally, with an impact on the world economy. To deal with these issues, smart agriculture solutions are evolving that combine the Internet of Things and machine learning for early disease detection and control. Many such systems use vision-based machine learning methods for real-time disease detection and diagnosis. With the advancement in deep learning techniques, new methods have emerged that employ convolutional neural networks for plant disease detection and identification. Another trend in vision-based deep learning is the use of vision transformers, which have proved to be powerful models for classification and other problems. However, vision transformers have rarely been investigated for plant pathology applications. In this study, a Vision Transformer enabled Convolutional Neural Network model called "PlantXViT" is proposed for plant disease identification. The proposed model combines the capabilities of traditional convolutional neural networks with the Vision Transformers to efficiently identify a large number of plant diseases for several crops. The proposed model has a lightweight structure with only 0.8 million trainable parameters, which makes it suitable for IoT-based smart agriculture services. The performance of PlantXViT is evaluated on five publicly available datasets. The proposed PlantXViT network performs better than five state-of-the-art methods on all five datasets. The average accuracy for recognising plant diseases is shown to exceed 93.55%, 92.59%, and 98.33% on Apple, Maize, and Rice datasets, respectively, even under challenging background conditions. The efficiency in terms of explainability of the proposed model is evaluated using gradient-weighted class activation maps and Local Interpretable Model Agnostic Explanation.
Discriminative Kernel Convolution Network for Multi-Label Ophthalmic Disease Detection on Imbalanced Fundus Image Dataset
Jul 16, 2022



Abstract:It is feasible to recognize the presence and seriousness of eye disease by investigating the progressions in retinal biological structure. Fundus examination is a diagnostic procedure to examine the biological structure and anomaly of the eye. Ophthalmic diseases like glaucoma, diabetic retinopathy, and cataract are the main reason for visual impairment around the world. Ocular Disease Intelligent Recognition (ODIR-5K) is a benchmark structured fundus image dataset utilized by researchers for multi-label multi-disease classification of fundus images. This work presents a discriminative kernel convolution network (DKCNet), which explores discriminative region-wise features without adding extra computational cost. DKCNet is composed of an attention block followed by a squeeze and excitation (SE) block. The attention block takes features from the backbone network and generates discriminative feature attention maps. The SE block takes the discriminative feature maps and improves channel interdependencies. Better performance of DKCNet is observed with InceptionResnet backbone network for multi-label classification of ODIR-5K fundus images with 96.08 AUC, 94.28 F1-score and 0.81 kappa score. The proposed method splits the common target label for an eye pair based on the diagnostic keyword. Based on these labels oversampling and undersampling is done to resolve class imbalance. To check the biasness of proposed model towards training data, the model trained on ODIR dataset is tested on three publicly available benchmark datasets. It is found to give good performance on completely unseen fundus images also.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge